“Allelic dropout in the PCR is a condition in which one or both the alleles in the heterozygous condition fail to amplify and cause a fault in results. In this article of our series PCR troubleshooting, I will explain what the allelic dropout is, and how it occurs with possible troubleshooting.”
Polymerase Chain Reaction is an important genetic tool that has been utilized to study genetic disease, and gene variations and used in downstream applications like DNA sequencing or microarray. The main function of PCR is to generate copies of DNA. However, it has been increasingly used in genotyping and disease studies.
Related article: 6 Optimizations to Improve PCR Genotyping.
PCR is a highly sensitive technique that needs an expert hand and eyesight to perform and interpret results, respectively. However, not only for beginners, it is a frustrating and tedious task for experts as well to get conclusive results, all the time.
No amplification, non-specific amplification, primer-dimers and allelic dropout are common problems everyone faces during their PCR venture. It becomes so crucial to avoid and/or overcome such problems when we have to deal with disease samples and perform sequencing.
So far, previously in this serious- in PCR troubleshooting 101 we have discussed the non-specific amplification. Continuing this series, in this article, I will discuss an important and rare condition ADO (allelic dropout).
Whether you are a seasoned expert with immense field experience or just a beginner in the field, understanding allelic dropout and how to deal with it, is crucial for getting excellent results in your PCR.
Read the previous article: PCR Troubleshooting 101: How to Address Non-specific Amplification.
Stay tuned.
Key Topics:
What is an allelic dropout in PCR?

ADO is referred to as failure amplifying target allele in PCR. In 1991, Navidi & Arnhein explained the concept of ‘partial amplification failure’ Which was later on in 1997, explained comprehensively by Lissens and Sermon. The exact definition of ADO is given here.
“When a PCR reaction fails to amplify one or both the heterozygous alleles due to either mutation or suboptimal PCR conditions and lead homozygous misinterpretation is known as an allelic dropout in PCR.”
If you find it difficult to understand, let’s take an example to make it easy for you. Let’s say you are trying to identify a single gene disorder having any mutation (using a PCR obviously!). You have a ‘defined’ protocol for that particular mutation. You also have set strict validation rules using positive, negative and internal controls.
Now you run the PCR and load the results. You observed single or no bands in the heterozygous condition. What does it show? Both homozygous wild and mutant alleles are amplified (in individual reactions) but how do the heterozygous fail to amplify?
Is it possible, technically? No. This is the case of allelic dropout. It shows that the target sequence may get any unknown mutation and resultantly, limited primers to amplify it. Or our PCR conditions aren’t optimal to amplify it.
The absence of both the heterozygous bands can easily be understood and ruled out, but when it comes to a single allele– a single allele from the heterozygous is a dropout, the case becomes very difficult as it clearly misinterprets the homozygous results.
“The sample can easily be misinterpreted as homozygous for the given mutation or allele if only a single allele dropout.” Lissens and Sermon (1997) during their study on preimplantation genetics on cystic fibrosis, identified a 25% population of mutant cells having allele dropout.
The present condition is often encountered during DNA or genetic testing while doing PCR for highly polymorphic regions STRs or VNTRs. Take a look at the example given below.
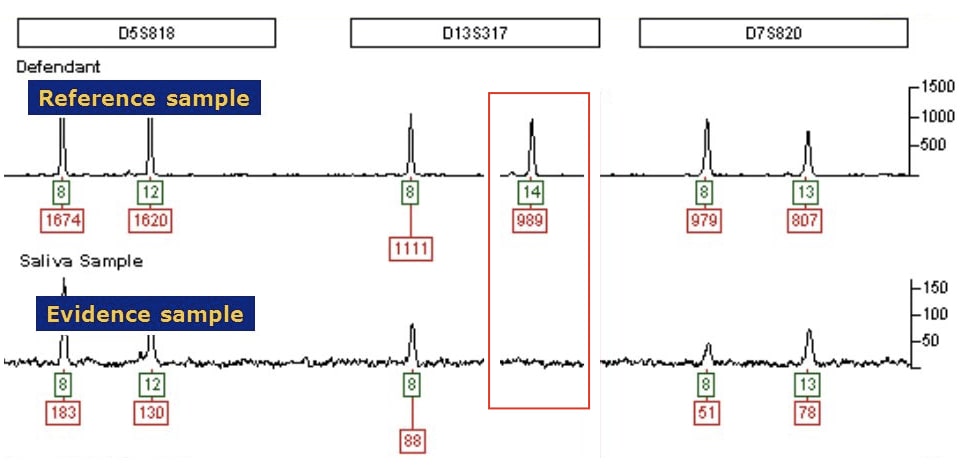
So how does it occur?
Causes for ADO:
The main cause of ADO is the failure of primers to anneal and amplify the target location either by sequence alteration in the target sequence or by suboptimal PCR conditions. Let’s understand each scenario, one after another.
Genetic alteration: Sequence variation or mutation in any of the alleles of the target sequence cause allelic dropout. Importantly, oftentimes, if single or two-base alterations are present in the middle of the sequence the primer can somehow manage to amplify the target.
However, the chances of ADO becoming dramatically increased, if the alteration may be located on the 3’ or 5’ end of the primer. In this case, the primers can’t anneal with the target location. This particularly becomes problematic for populations with high genetic diversity and while targeting highly variable genomic regions.
Suboptimal PCR conditions: inadequate annealing temperature, suboptimal primer concentration and PCR condition, non-validated PCR protocol and other factors involved in the reaction preparation are common causes of ADO.
Low-quality DNA: low quality and quantity DNA can also cause PCR inhibition. Contaminated template DNA reduces the efficiency of primer binding, and activity of Taq DNA polymerase and blocks the site for amplification.
What problems does it cause?
It’s certainly difficult for a newbie student or unrecognized scientist to understand the allelic dropout and they can easily misinterpret the results. ADO is totally a different scenario than no-amplification and non-specific amplification.
Here, one of the heterozygous alleles fails to amplify. So if the normal allele fails to amplify, the results are wrongly manifested as homozygous normal and vice versa for the mutant allele. The illustration is given in the image below.
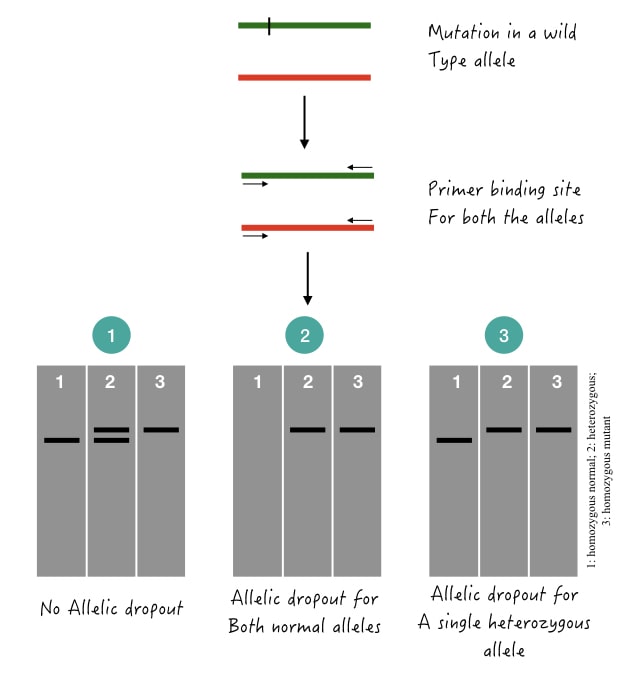
If both alleles are dropouts, it clearly indicates the absence of that particular genotype from a sample, individual or population. Now how to overcome such problems or perform troubleshooting?
ADO troubleshooting:
In this section, I will explain some common troubleshooting and some specialized troubleshooting from my personal experience.
Primer design and concentration: Design and choose both the forward and reverse primers so carefully. Both primers should have a nearly similar or adequate melting temperature and can 100% complement the target location. Avoid locating your primers in the highly variable region.
Use an adequate concentration of primers to amplify the target. Perform gradient PCR to optimize the concentration of primers. Higher primer concentration may result in non-specific amplification and primer dimers.
In silico PCR: validate your primer binding efficiency, target specificity and amplification accuracy by doing in silico PCR analysis. Ensure that the primer set should have an equal capacity to anneal and amplify both wild-type and mutant allele.
Target multiple loci: The probability of ADO and misinterpretation of results can greatly be reduced by following multiplex PCR. Target multiple loci of the same target sequence and check if any loci fail to amplify or not. This ensures results guarantee many folds.
Nested PCR: Yet another proven and safest way to avoid ADO is doing nested PCR. Design two sets of primers in which the second set of primers is nested to the first set. Any ADO can be resolved by following the results pattern of nested PCR.
Related article: What is nested PCR?
PCR optimizations: Optimization or re-evaluating your PCR conditions can also help overcome ADO. Adjust your annealing temperature, Taq DNA polymerase concentration, PCR buffer, template DNA concentration, etc.
High-fidelity Taq DNA polymerase: The low-quality Taq DNA polymerase sometimes creates problems in amplification, although it’s not one of the main reasons for ADO but, by using the high-fidelity DNA polymerase the chances of allelic dropout can be reduced.
Use PCR controls: Use positive, negative and internal reaction controls. Controls in PCR give us the flexibility to identify the ADO and understand the results.
DNA sequencing: If the allelic dropout is diagnosed, sequencing can help to understand why it occurred. Sequence the allele in order to understand if any new or pre-existing sequence variation harbors the ADO.
Other: Use high-quality template DNA and avoid any contamination during PCR reaction preparation.
Related article: Polymerase Chain Reaction (PCR)- Definition, Principle, Steps, Procedure, Protocol, Applications and Types.
Wrapping up:
In conclusion, ADO isn’t a much bigger problem in the PCR, however, it’s important to know it in order to avoid misinterpretation of results. It, although, can be a major hurdle in disease-related PCR and preparing amplicons for sequencing.
It majorly causes problems in the interpretation of homozygous genotypes thus results must be carefully evaluated and a second opinion from an expert can be considered before delivering the results.
I hope you like this article. Please share and bookmark the page.