“PCR genotyping is a technique to identify pair of alleles present in a sample thereby the condition- homozygous dominant, homozygous recessive or heterozygous.”
Thalassemia and sickle cell anemia-like conditions are very rare and hence oftentimes I found PCR genotyping results as either thalassemia minor or normal.
But when it comes to thalassemia major, the condition becomes complicated. “PCR genotyping demonstrates that your baby has genotype for Thalassemia major for mutation IVS1-5 of Beta-globin gene.” I had given this statement many times. during my work in the prenatal lab, But I actually hate to say this!
To further, I just say to the couple that you should go for an abortion. Quite a heartbreaking situation for a couple and me too. I’m a molecular geneticist and have decent working experience in genotyping other techniques.
Genotype is a genetic variant of a gene whilst genotyping is a process to determine or study it. For example, genotypes are denoted BB, Bb, bb for a particular trait or gene. Genotyping techniques are SNP genotyping, indel genotyping, PCR genotyping and genotyping by sequencing.
Genotyping and PCR genotyping are terms commonly used for DNA tests and in research. Mouse genotyping by PCR or sequencing is a popular technique to study a specific trait, gene or phenotype.
Sometimes if you came across a DNA test you may hear that your PCR genotyping is homozygous or heterozygous for this or that particular gene, disease or trait. So what exactly a PCR genotyping is! That’s what the topic of this blog is.
In this article, I will explain the genotyping by PCR technique and will give you 6 optimization options to improve it.
Stay tuned.
Related article:
Key Topics:
PCR genotyping- explained in a Lyman:
PCR genotyping is a technique often employs to determine genetic variance for a gene, trait, disease or phenotype. Such a technique can tell us if the genotype is homozygous, heterozygous, dominant or recessive.
Genetic literature describes two common genotypes for a gene as homozygous and heterozygous with a similar type of two alleles and two different alleles, respectively.
Two similar types of alleles produce two conditions viz homozygous dominant and homozygous recessive having two same types of dominant allele and two same types of the recessive allele, respectively.
Depending upon these conditions a genetic disease is also characterized. So when someone says that the person has a homozygous dominant genotype for a disease, meaning it has two dominant alleles for the mutant gene and will get the disease.
Vice versa other conditions are defined. Here is the table that makes you understand more.
| If allele “B” is mutant and allele “b” is normal | |||
| Homozygous | Dominant | BB | Disease |
| Recessive | bb | Normal | |
| Heterozygous | Bb | Disease |
| If allele “B” is Normal and allele “b” is Mutant | |||
| Homozygous | Dominant | BB | Normal |
| Recessive | bb | Disease | |
| Heterozygous | Bb | Carrier |
PCR as a tool for genotyping has lucrative utilities in genetic research and diagnostics. It has a pivotal role in disease diagnosis too, however, it uses only available information to determine the genotype.
Monogenic diseases, like thalassemia, sickle cell anemia or cystic fibrosis can be ruled out by PCR genotyping technique, qualitatively and quantitatively. It has significant value as a validation tool in the plant, animal or microbial genetic research.
Traits, phenotypes or disease governing genotypes, transgene and transgenic mice, recombinant gene and plasmid insert can be genotyped by the present technique.
The conventional PCR genotyping method can give us information regarding the presence or absence of a particular genotype in the sample, while, qPCR determines the amount of that particular genotype in a selected tissue sample.
In gene expression-based research and transgenic mice studies, genotyping by qPCR provides knowledge of whether or not a gene or transgene is expressed.
Genotyping by either PCR technique is a rapid, cost-effective, accurate, sensitive and reproducible testing option and thus getting roars day by day.
It can also genotype microbial genes. But this state-of-the-art technique has some serious limitations. It finds difficulties in distinguishing genotypes.
When it comes to SNP genotyping or genotyping for small ‘indels’, it is very difficult to differentiate differences in the genotyping pattern. So newbies and inexperienced lab personnel find it difficult to genotype by PCR.
Furthermore, preparing reactions is often difficult and may get cross-contamination.
In the upcoming section, I’m giving you 6 optimization options to improve the PCR genotyping.
Read more: Differences between PCR vs qPCR
6 Optimizations for PCR genotyping
- Design separate primers that produce different lengthed products
- Prepare separate PCR reactions
- Avoid cross-contamination
- Prepare a single multiplex reaction
- Use PCR controls
- Run optimizations in a gradient
Design Separate primers that produce different lengthed products:
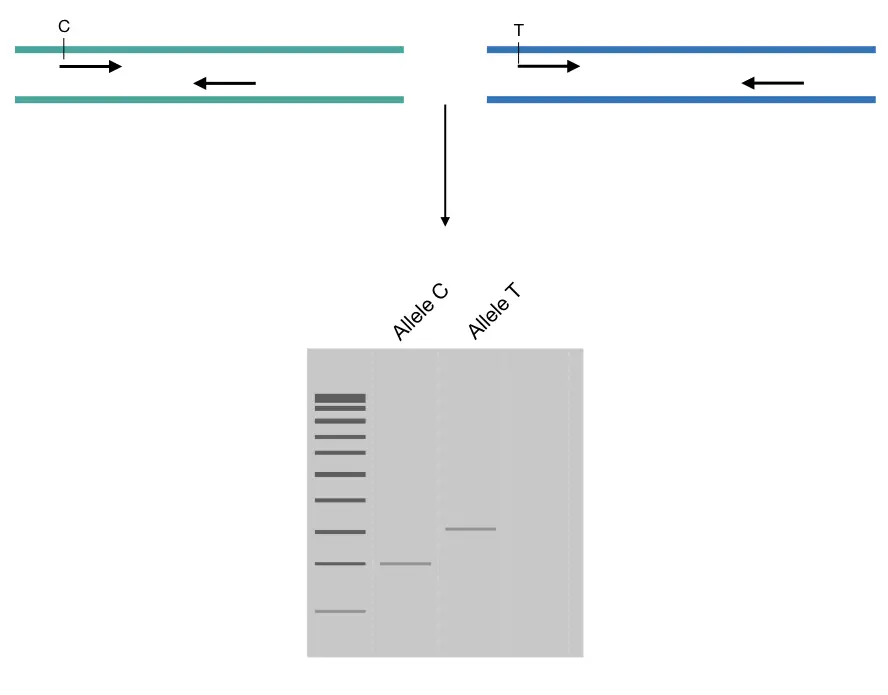
The common and most approachable way to perform the genotyping by conventional PCR is by altering the length of primers or primers in a way that produces differences in the size of final amplicons. See, we are distinguishing wild type vs mutant or norma vs transgene, which perhaps has SNP or small ‘indel’. Meaning alleles for such genes aren’t separable on a gel.
Gel electrophoresis separates DNA according to its size, but in the case of SNP, no such size difference occurs, furthermore, smaller deletions or insertions of 10 to 15nt also can’t separate well to interpret the results.
When we design primers that can amplify alleles but produce different lengths for the normal and the mutant allele, we can separate them on a gel and interpret the results.
For example, for C>T transition, one primer amplifies the allele with C and gives a 220bp band while another primer set amplifies the T allele and gives a band of 340bp. Such results are easy to interpret on a gel.
To optimize the PCR genotyping by size difference, we only need to adjust the binding site of only one primer of either set. Usually, alteration in the annealing site of the reverse primers makes the assay easy.
Note that prepare separate reaction, and run on a gel in a separate lane. Take a look at the image.
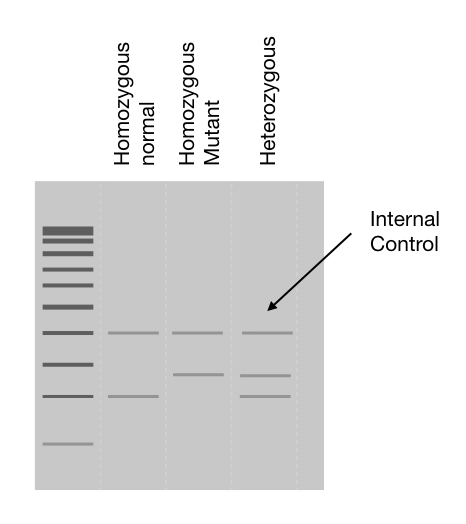
Prepare a separate PCR reaction:
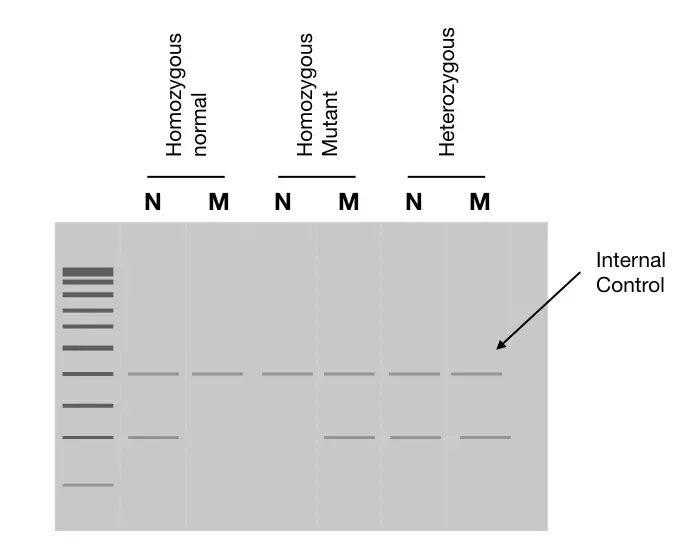
Yet another conventional, handy and easy way for PCR genotyping is to prepare a separate reaction for each allele. i.e. one reaction for normal allele and another for mutant.
Run each reaction for a specific allele in a separate lane, the presence or absence of band shows which allele is present. Take a look at the image. The image depicts that the single band in a normal is a homozygous normal and a single band in the lane mutant is a homozygous mutant. In contrast, two separate bands in two separate lanes for normal and mutant show heterozygous conditions.
Preparing and running PCR reactions separately is an easy, rapid and more sensitive approach. Although it needs a dedicated internal PCR control to interpret the results.
10 secrets to Prepare a PCR reaction.
Use PCR controls:
The PCR control system supports the accurate interpretation of results. Suppose if we get amplification for the normal allele but not for mutant, how can we say that if the allele is absent or the personnel hasn’t prepared the reaction well?
Means, we might forget to add something, right! It happens. So we need internal control. Which runs parallel to our target allele. Internal control is added in the reaction which amplifies a different region than our target but gives uniform DNA bands in all reactions.
This concludes that the reaction has all the ingredients and also works finely. Meaning, even if we don’t get any of the target allele bands (mutant or normal), the presence of internal control bands shows that everything in the reaction is fine.
Further to this, also incorporate negative and positive controls to make the interpretation even more powerful. To know more about each PCR control, this article will help you,
Type of PCR Controls- Negative, Positive and Internal Controls.
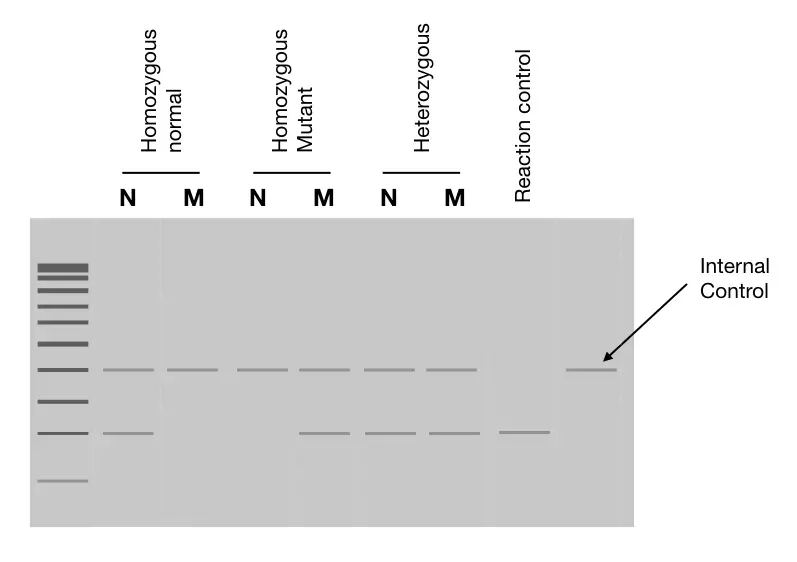
Prepare a single multiplex reaction:
Multiplexing is excellent as it saves time and reagents for a PCR reaction. Although the sensitivity is less. The reaction can be combined while dealing with the optimization to produce two different lengthed products.
All four primers along with all the PCR ingredients are run in a single reaction (with the control as well). In a gel, DNA bands are observed in a single reaction.
Note that, our goal is to combine the PCR for two separate alleles of a single genotype, not all the reaction. Take a look at the image below.
Avoid cross-contamination:
Working with many primers increases the risk of cross-contamination which is one of the most serious issues in any PCR reaction. Even though you can rule it out, you have to throw all the reagents, at last.
Here in PCR genotyping, our deal is with at least 4 to 6 different primers (One set for normal allele, one for mutant and one for control) so mix-up of any primers leads to cross-contamination and false-negative and false-positive results.
What to do?
- Prepare a unique working style for genotyping and a separate SOP.
- Throw tips, do not reuse it and take care that it should not be re-used, once used.
- Add Taq DNA polymerase separately or at the end to avoid chaos in the reaction.
- Autoclave all the autoclavable utilities and wash other utilities with 70% ethanol.
- Prepare reaction in a separate PCR workstation.
Run optimization in a gradient:
Increasing the number of ingredients in a PCR reaction also increases the complexity of the experiment. PCR is a very sensitive and high throughput technique that needs special care and high-end expertise.
Reaction failure and false (positive/negative) results are common while working with these much of ingredients. It is recommended to optimize the quantity of each ingredient, PCR cycling conditions and time required before using it for actual in vitro study.
Set various gradients for different primer concentrations, cycling conditions, annealing temperatures etc. Choose the best condition for your reaction. Perform many repetitive runs and use the one as a standard SOP.
Wrapping:
Genotyping has now become advanced, techniques like microarray or sequencing can identify thousands of genotypes in a single run, however, techniques like PCR genotyping also have significance in this scenario.
Such techniques are cost-effective, rapid and reduce complexity and thus have great value in the diagnosis of monogenetic disease in prenatal, post-natal and preimplantation genetics.