“Primer dimers are short amplified products formed by the binding of primers during PCR amplification. Let’s learn what primer dimers are in PCR, qPCR, gel electrophoresis and DNA sequencing.”
PCR (Polymerase Chain Reaction) is a core and important technique in molecular biology. Its function is to amplify the DNA, which means, generating copies of DNA. However, during PCR, we have to encounter various challenges.
The primer dimer is one such PCR problem that commonly occurs in almost every PCR reaction. It doesn’t interfere directly but can mislead the results. So from the very first day of your PCR journey, you should know about it.
This article is an introductory article on primer dimers. If you want to learn about some advanced troubleshooting on how to address Primer dimers, you can click the link and read our article.
Key Topics:
What is a Primer dimer?
Primer dimers are short, unwanted and unintended PCR products caused by the amplification of primers.
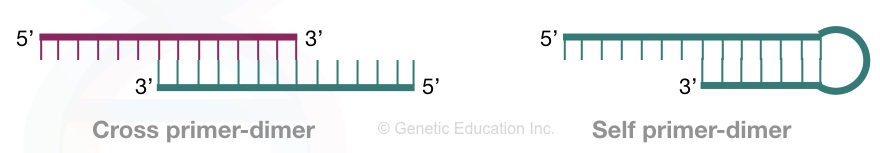
Two types of primer dimers are reported commonly in PCR reactions– Self primer (Homodimer) dimer and Cross primer dimer (Heterodimer).
Homodimer:
Homodimers are self-primer dimers formed when identical primers bind to each other. In this case, the complementarity between the identical primers is involved in dimer formation and synthesis.
Notedly, homodimers interfere directly with the target amplification as one type of primer is not available for amplification. PCR reaction without any target amplification and with high primer dimers confirms the presence of homodimers.
To overcome it, carefully look at each of your two primers and check for complementary nucleotides. Identify it and replace the primer with another set of primers or a single primer.
Heterodimers:
Heterodimers are cross-primer dimers formed when two different primers bind with each other. In this case, the forward and reverse primers share some of the regions or nucleotides, bind and amplify in the PCR reaction.
Heterodimers also interfere with the target amplification by reducing the amount of amplification. A reaction with fever amplification product and high primer dimer confirms the presence of heterodimers.
To overcome it, carefully look at your primer sequence and change it with another primer set.
How primer dimers are formed?
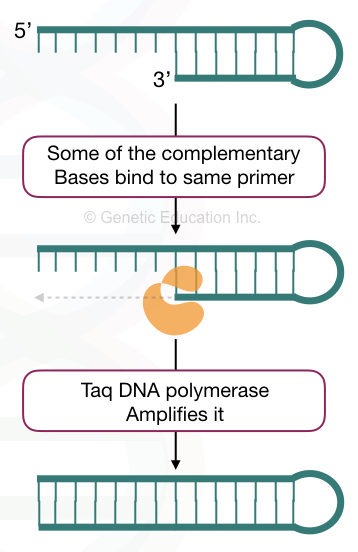
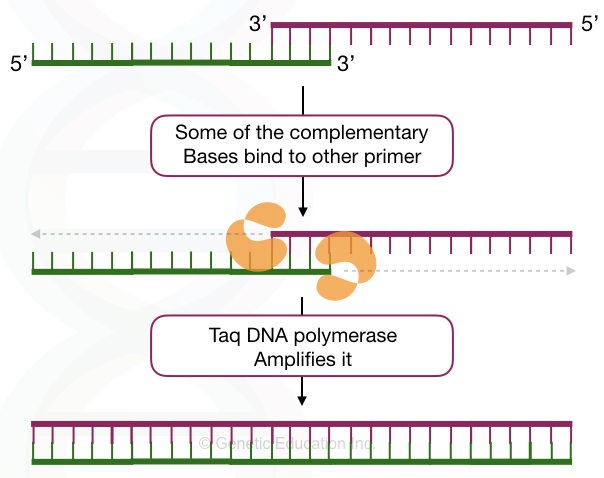
Dimers in PCR are formed when the primers have complementary nucleotides. Understand it by a simple explanation. In normal physiological conditions, DNA remains in the double-stranded form, if they have complementary nucleotides.
Primers are also, sort of, DNA sequences. So when they find any complementary region on the same type of primer or another primer, they bind. This can also occur early during the reaction preparation.
During the amplification step, when Taq DNA polymerase synthesizes or amplifies the target, such unintended short structure is also amplified and shows their presence in the results.
Studies also showed that the Taq DNA polymerase-governed amplification has also been started early during the PCR reaction preparation step as the Taq also shows some activity at room temperature.
So the dimerization process is actually initiated during the reaction preparation and continued till the end of PCR. Thus, it’s advisable to add either primers or Taq DNA polymerase at the very end of the reaction.
Primer dimers in PCR:
Primer dimers in the PCR, as I said, occurred by many reasons (I have explained each reason in another article, the link is given above). However, it can be identified using agarose gel electrophoresis.
It can be ruled out by following various strategies which are also mentioned in the same article, the link is given above.
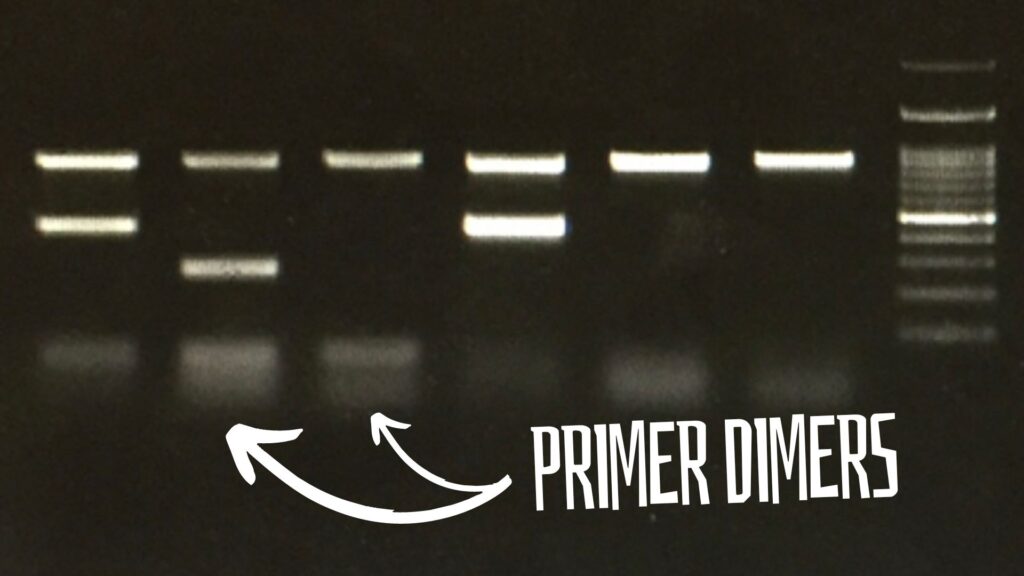
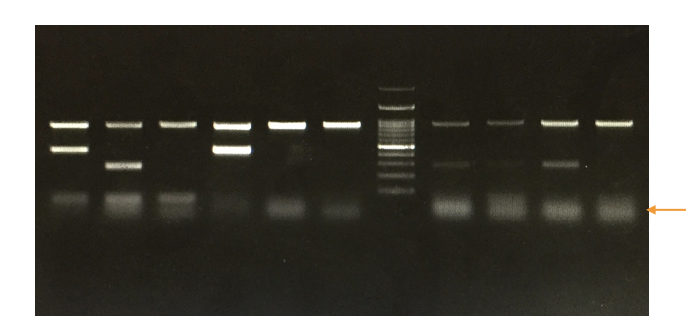
Primer dimers in a gel:
In a gel, the primer dimers appear at the very end of the positive side. Dimers are very short and unintended amplification products. So, it appears at the very end of the positive side with diffusion.
Technically, the dimer bands look blurred and unsharp as it diffuses in the gel very fast. Usually, they appear near the 20 to 50 bp zone.
Primer dimers in qPCR:
Dimers can also interfere with the quantitative PCR reaction much like the native (conventional) PCR, however, can be identified differently. Broadly, qPCR relies on the emission of fluorescence signals for results and interpretation.
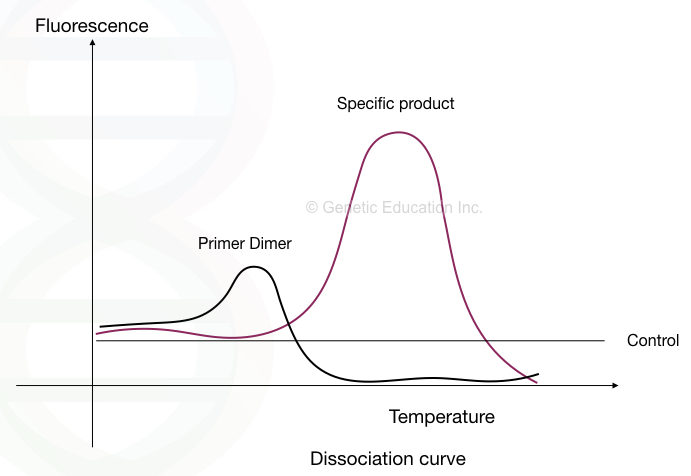
Dimers in the qPCR also amplify and emit fluorescence. Also, it amplifies as short products at a rapid pace and thus appears earlier to the main target amplicon in the amplification plot. The plot height is also very short.
To validate and rule out primer dimers, a melting curve analysis has been carried out. By gradually increasing the melting temperature the main target amplicon, non-specific amplicon and primer dimers are identified.
See the image below.
Primers dimers in DNA sequencing:
Primer dimers also appear in the DNA sequencing process and can mislead the sequencing results. In sequencing results, it appears as a background amplification or noise during the reading process.
Related article: DNA Sequencing: History, Steps, Methods, Applications and Limitations.
Wrapping up:
Dimers in PCR or any other reaction (qPCR, RT-PCR, nested PCR or DNA sequencing) are a known and common problem. However, it can be ruled out by taking care of several factors. I am giving you the link for the troubleshooting article at the end of the article that you can read.
If you would like to learn more about PCR and related topics, you can subscribe to our blog or read more articles here.
Related article: PCR Troubleshooting 103: How to Address Primer-Dimers.
Sources:
Brownie, J et al. “The elimination of primer-dimer accumulation in PCR.” Nucleic acids research vol. 25,16 (1997): 3235-41.