“The chemically synthesized exact copy of the genome of an organism is called synthetic genome which is used to manipulate phenotypes or to create a new life form.”
Synthetic genome is a science of redesigning and synthesizing entirely new DNA sequences. It’s a fascinating thing! looks easy but it’s not.
Actually, we are here copying the genome or trying to write it as it is, in its original form. We are not writing or designing a new DNA sequence. And it’s quite impossible, currently.
Let us understand it with an example.
It is just like replacing a car engine, instead of replacing a particular part, we are replacing our car engine to boost the performance, however, the body of the car is still the old one.
Now imagine the scenario with a cell.
In the process, we will insert a synthetically designed, exactly identical and artificial genome into a target cell. Here we are introducing a new genome by replacing the native one in order to boost the performance of a cell.
Understand the mechanism, we are not manipulating the pre-existing genome, that’s totally a different thing and covered as gene editing.
So you may wonder why we are doing it? In the present article, I am discussing exactly that. I will make you understand what a synthetic genome is and why we do it. Alongside, I will also explain several techniques and other relevant information.
Stay tuned,
Related article: The Human Genome Project: Aims, Objectives, Techniques and Outcomes.
Key Topics:
What is a synthetic genome?
From the J. Craig Venture Institute, Rockville, Maryland, scientist Daniel Gibson and co-workers had first introduced the concept of a “synthetic genome.”
They had used three different species from which one’s genome was copied, assembled in another and expressed in some other organism.
They had selected Mycoplasma mycoides and synthesized 1.1 million base pairs of its entire genome, assembled it in yeast cells and inserted it into the Mycoplasma capricolum.
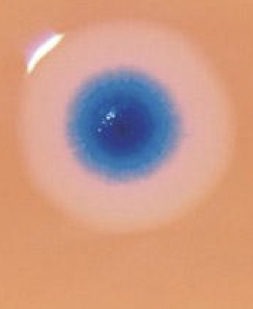
With it, to validate results they had inserted a fluorescent marker gene. After several cell divisions, blue-colored colonies were observed indicated the success of the experiment.
The artificially tailored genome of Mycoplasma mycoides was copied, some non-essential genes were removed and fever modification was made in the target organism to express the entire genome.
It is also noticeable that they were unable to do some complicated modifications such as mimicking DNA methylation patterns.
DNA methylation has been associated with gene expression can’t be copied exactly. It’s nearly impossible to locate every methyl group like the native sequence.
For overcoming this problem scientists had removed the enzyme that degrades the unmethylation sequences. What was done by Gibson D and co-workers was truly incredible, and opened new doors for new genomic research.
However, it’s also important to note that without the help of recombinant DNA technology, they couldn’t do this.
The recombinant DNA technology was evolved during ‘70; restriction endonucleases, ligases and other essential tools and techniques developed during this period.
Using the endonucleases and ligases scientists were able to cut and seal the DNA which enable them to construct artificial DNA parts.
Restriction endonucleases: these are the enzymes that cut or digest the DNA sequence at its specific location called the recognition site. The entire process is called restriction digestion.
Related article: Restriction digestion.
Ligase: ligase is the class of enzyme used to join or seal the DNA sequences, it forms a phosphodiester bond between two adjacent nucleotides and seals the gap.
Related article: Ligase.
Definition:
“Re-designing, copying and synthesizing the entire genome or all DNA sequences of an organism is called the synthetic genome.”
Or
“Artificially created genome used in order to create new life is called synthetic genome and the science is called synthetic genomics.”
Methods for constructing a synthetic genome:
Polymerase cycling assembly:
The polymerase chain reaction is one of the best methods to perform in vitro replication, it has the power to copy the entire strand of DNA or gene of our interest. However, it can only amplify the DNA of several thousand base pairs.
Using the cycling assembly method combined with the polymerase chain reaction, we can synthesize a genome of more than several kilobase pairs, one of the examples is the Phi X 174 virus- the entire genome of this virus is synthesized using polymerase cycling assembling.
In the process, 40 to 60 nucleotide long oligonucleotides complementary to our DNA are selected. The unique design of the oligos enables the synthesis of an entire genome as the 20 nucleotide sequences on both ends of each oligo binds to two different complementary to the opposite strand.
- In the very first step the denaturation is done which separates two DNA strands.
- Hybridization of DNA sequences is done at 60°C followed by the polymerization through the Taq DNA polymerase.
- All the gaps between all the DNA fragments are sealed by ligase.
Read more on PCR: Polymerase chain reaction.
TAR- technology:
TAR- Transformation Association Recombination method is used to achieve homologous recombination between plasmid and genomic DNA.
The yeast artificial chromosome- YAC vector is constructed in such a way that behaves like the organism’s own chromosome. Using the polymerase chain reaction the gaps are repaired between the fragments utilizing the extension primers.
(Note: extension primers are the set of primers used to bind sequences flanking to our gene of interest).
The process of homologous recombination occurs between the YAC vector with the DNA cassettes and the yeast’s own chromosome. A DNA cassette is inserted into the yeast’s own chromosome. And replicates there.
Gibson assembly method:
The present method was originally developed by Gibson and co-workers and applied in the synthesis of the world’s first synthetic genome.
The method is a single-step isothermal reaction that synthesizes larger DNA sequences than the polymerase cycling assembly method.
The isothermal reaction is performed followed by exonuclease and polymerization.
The exonuclease creates an overhang complementary sequence on both ends and hybridizes.
Then in the next step, the special type of DNA polymerase expands the DNA sequence.
In the final step, the ligase seals the gaps.
The Gibson assembly method can synthesize the genome of more than 6Kb.
These are not fully functional techniques but are several ways like “trial and error” to synthesize an artificial genome and to achieve efficiency.
Note
Gibson and co-workers failed in their 99% of experiments for synthesizing the genome.
Synthetic genome technology is used to create some of the economically important microbes such as biofuel secreting microbes, alcohol-producing microbes, for decontaminating toxic waste and to track down the tumor cells.
Usually, the process completes in five steps.
Writing or analyzing the entire genomic DNA sequence of the target organism.
In the very first, steps, scientists sequence the target genome (which they wish to synthesize), extract sequences, analyze computationally and collect information like
- Order of sequences
- Structure of the genome
- How sequences are located on chromosomes
- Structural hierarchy of every gene
- Methylation patterns, essential genes, histone modifications and other epigenetic factors.
Collectively, it’s referred to as pre-processing.
Synthesizing the genome using one of the methods listed above:
I have explained popular techniques for the purpose so not discussing that part here, you can go ahead and check it out. In this step, the selected genome is synthesized in fragments.
The reason for it is that we do not have that much powerful technology to process the whole genome in one run or experiment.
Every fragment is prepared in one or different laboratories. For example, BioFab like groups are providing promoters and other regulatory genetic elements required to construct a synthetic genome. BioFab currently has more than 350 promoters that are commercially available.
Assembling the genomic fragments:
Researchers collect every single fragment prepared in separate laboratories and join them as per the original sequence. That’s a bit tedious job, actually. Enzyme ligase is applied to connect every fragment.
Transplanting the synthetic genome in the target organism:
Incorporating or transplanting artificially prepared genomes into a host cell is yet another challenging task and is tougher than gene therapy or gene editing. The reason is that here we are not only transplanting the genome but also removing the original one.
Viral and non-viral vector-mediated approaches are used here. Usually, smaller sequences are transplanted using a plasmid. It is also important to note that cells’ immune response may reject the insertion much like the graft rejection process.
Boot up the cell to express the genomic content:
“Booting up” our synthetic genome is also one of the challenges as we don’t know more about how the entire mechanism is actually working.
Scientists must have to do some modifications to achieve the expression, for example, removing enzymes that cleave the unmethylated DNA sequences or removing those sequences which are not essential.
Application of synthetic genome:
- Artificial genome synthesis can be beneficial in many ways, however, Gibson and c-workers (original developers of the concept) are still not clear about how it is useful to us. Nonetheless, some of the possible applications of it are enlisted here.
- Using synthetic genome technology, the newer biosynthetic pathway can be incorporated in microorganisms to produce biofuels and alcohol.
- It can also be utilized as a detoxification agent.
- Further to this, humanized antibodies can be generated artificially.
- The important aspect of the present technique is to produce minimal artificial genomes of model organisms with the minimal genome. Unnecessary or useless sequences can be removed.
- Humanized pig or animal models can be developed to create artificial organs which can solve the problem of transplantation failure.
- Newer metabolic pathways can be inserted into the target organism to fulfill new functional requirements.
- Other than this, it might have some therapeutic applications for human health. However, these are some of the possible uses, none of the synthetic genome models is now fully available for therapeutic uses yet.
Put simply, we can prepare microbes just like robots, equipped with functions only helpful to us.
Limitations of the synthetic genome:
- Developing an entire synthetic genome itself is a challenging, tedious and time-consuming process and yet success isn’t guaranteed.
- It’s nearly impossible (as of now) to construct the whole genome in a single laboratory. Libraries of sequences are synthesized in separate laboratories which elevates the chances of error.
- The overall cost of the project is yet another factor that is almost 10 times more than gene therapy.
- Booting up the artificially synthesized genome is another major problem, scientists still do not have knowledge about how exactly cell machinery works to boost up a cell.
- In addition to this, scientists have to modify a cell to adopt a new genome, even though it is exactly the same as its native one, the chance of rejection is very high.
- None of the present technology can synthesize exactly the same genome or an entire genome.
- Even though some of the bacterial and viral genomes are synthesized artificially, designing an entire genome of a eukaryote is just like a dream for now.
Recent trends:
The time when there have been controversies around the technology and lost interest in genomics gained scientists’ attention during the COVID conditions. We all know how dangerous corona is!
The highly contagious property and lethal consequences restrict scientific research around COVID. However, synthetic genomics opened doors for new research. Companies like “CODEXDNA” offer identical synthetic genomes of recent SARS-CoV-2.
On a technical note, they provide full genome, promoters, mouse-adapted genome, replicons and reporters which allow scientists to research without fear.
Conclusion:
Synthetic genome is actually a fascinating and interesting concept but for now, it is impossible to commercialize it. The process is time-consuming, the success rate is unpredictable, the cost is very high, the process is uncertain and the outcomes are unknown.
We scientists have to work hard to make it real and applicable to mankind.
Sources:
Baker, M. The next step for the synthetic genome. Nature 473, 403–408 (2011).


