“Operon is the cluster of genes expressed together from one single promoter. trp and lac operon are common systems found in bacteria.”
Frankly speaking, the concept of operon is a bit scary for students. Pupils don’t want operon to be asked in exams. The reason is that they feel it is complicated to understand. And perhaps it is! However, it isn’t hard enough to understand.
The concept of operon is associated with gene regulation in bacteria. It is different from eukaryotic gene regulation.
So the first key topic we will understand is how gene regulation occurs in bacteria? However, not all bacteria or bacterial genes obey the operon model. Some genes also follow non-operon models.
So now it is clear that for understanding the present concept we first have to know about the gene expression and regulation.
“Genes form proteins”, we know it, right! but with it, some genes also regulate their expression. In which amount, genes expressed in one particular type of cell are known as gene expression.
Let’s take an example, fat metabolised in a liver. The liver cell needs a high amount of fat metabolite enzymes and hence genes for it as well. But those genes aren’t very important in eye cells or skin cells or any other cell types. What do you think?
This mechanism is known as gene expression.
It’s a kind of “on” and “off” switch for a gene. Prokaryotes also follow the mechanism of gene regulation, like eukaryotes, the process is somewhat different, although.
In the present article, we dive into the world of bacterial gene regulation. Also, we are discussing trp and lac operon in this article. We will try hard to make you understand what the operon is!
Related article: What Is a Gene?- Definition, Structure And Function
So let’s start,
Key Topics:
What is operon?
In a bacterial operon, a single promoter controls the expression and regulation of many genes.
The first bacterial operon was studied in 1960. Notably, it is also reported in bacteriophage as well. Operons are also observed in eukaryotes too but it’s transcribed into monocistronic mRNA, unlike bacteria.
Polycistronic mRNA is a common feature of a bacterial operon. Which means, a single mRNA transcript forms various proteins.
In 1965, Francois J, Andre ML and Jacques M were awarded Nobel Prize in Physiology and Medicine for describing lac operon in E. coli.
Note
Monocistronic mRNA- A single mRNA forms a single protein
Polycistronic mRNA- A single mRNA from an operon translates various proteins.
Unlike the eukaryotic genes, prokaryotic genes have only a few regulatory elements. However, promotor is a common feature observed in both.
Before understanding the mechanism of regulation, we have to understand some key terms or elements of an operon. These elements help in inducing or repressing gene expression.
Promoter– Prokaryotic promoter is simpler than eukaryotes. A promoter is a polynucleotide sequence that facilitates RNA polymerase binding. Unlike the eukaryotic promoter, it doesn’t have distinct proximal or core promoter regions.
It has simple non-protein coding sequences located upstream to the cluster of genes before the operon starts. See the image below,
Once the RNA polymerase binds to it, mRNA synthesis is initiated and it converts into mRNA transcript.
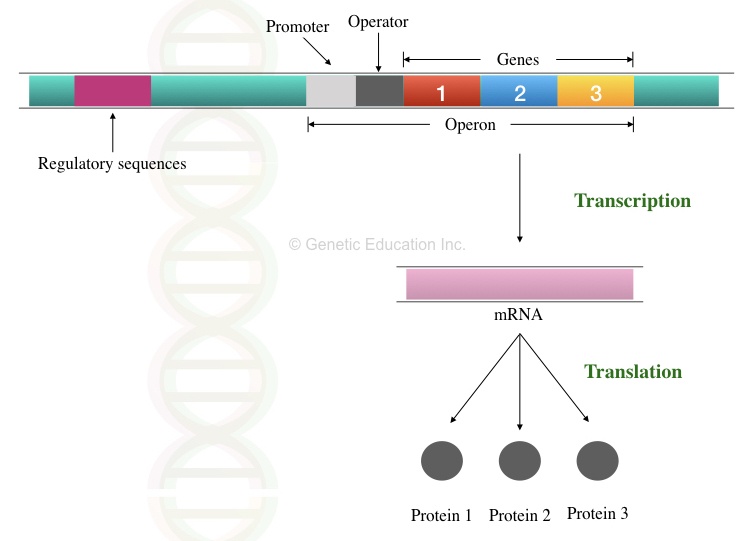
Operator– Also it’s a sequence of non-coding DNA at which either inducer or repressor binds. Usually, the operator is located downstream to the promoter. The activity of RNA polymerase is dependent on the operator.
Inducer– Inducers are smaller molecules or proteins that replace the repressor protein from the operator site for inducing gene expression. It induces transcription once bound to the operator.
Repressor– Also, repressors are smaller molecules or proteins that abort transcription. When the repressor links or attached to the operator sequences, RNA polymerase can’t recognise the promoter. Thus mRNA transcription missed.
However, the repressor itself can’t bind to the promoter because its confirmation needs to be changed. Another smaller molecule does do this function for it. The corepressors!
Corepressor– corepressors are also smaller molecules facilitating binding of the repressor to the promoter by changing its conformation.
As we know, the repressor can’t function properly, until and unless it’s shape changes. This function is performed by compressors.
Terminator– A type of nucleotide sequence ends the transcription is known as the terminator. These types of sequences are located at the end of the operon. Once RNA polymerase reaches there it’s detached from the operon.
These are the elements of the operon system. Understand it properly before going further.
Now let’s quickly move to the next section of our topic. Let us discuss trp operon.
Further read: microRNA (miRNA) and Gene Regulation
trp operon:
The E.coli operon system (a group of genes) that encodes enzymes for the tryptophan biosynthesis is known as trp operon. The entire mechanism of trp operon is regulated by the repressor mediated attenuation process.
The E. coli trp operon has 5 different genes. These are located one after another for encoding enzymes to facilitate tryptophan biosynthesis.
Like us, all other organisms also required proteins to survive and so E. coli too! Amino acids are the building blocks of life. They make proteins. A kind of amino acid tryptophan is an “essential amino acid” needed for bacteria.
First, let us understand the structure of the operon then we will understand how it works.
TrpE, TrpD, TrpC, TrpB and TrpA genes are located after the promoter region in the trp operon. The promoter has the RNA polymerase binding site and operator molecule to govern the entire process. See the image below,
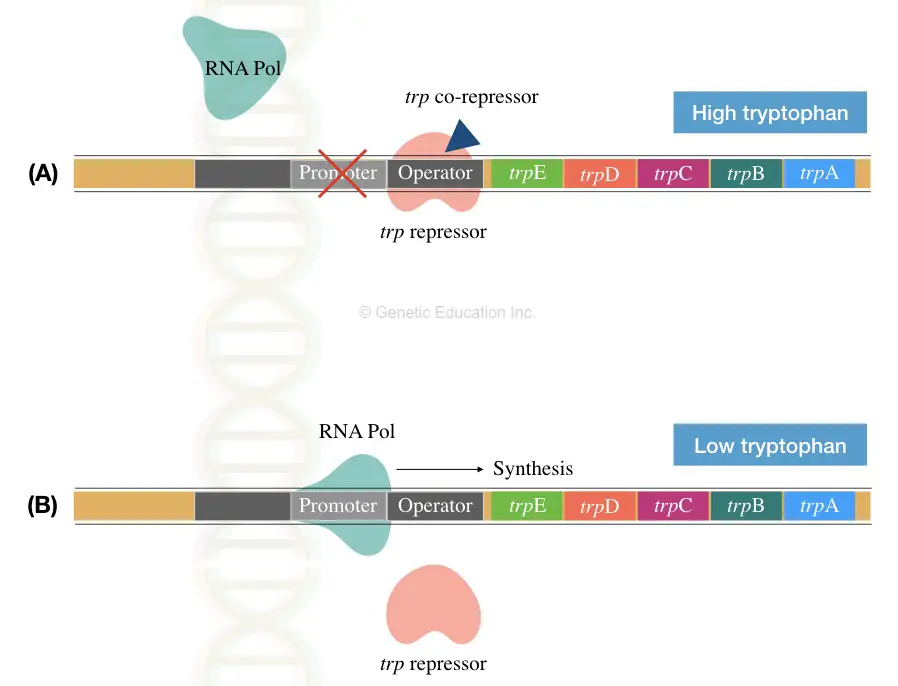
Upstream to the gene transcript, the operon also contains some of the important sequences known as leader transcript of 130 nucleotides (trpL). various genes functions are described here,
TrpE gene: Anthranilate synthase
TrpD gene: Helps the TrpE gene
TrpC: phosphoribosylathranilate isomerase
TrpB: Forms subunit of tryptophan synthase
TrpA: forms subunit of tryptophan
TrpR: forms repressor protein
As it is regulated by the repressor, it is also known as repressive operon as well.
Notably, all 5 genes transcribed into single mRNA but translate different proteins. And hence it is a polycistronic type of mRNA.
The entire mechanism of regulation is very interesting. Here in the present operon, the tryptophan itself works as a corepressor. It binds to the repressor molecule and changes its structure to facilitate binding with the activator.
The E.coli can take tryptophan from the environment as well as synthesise their own. Therefore, when a higher amount of tryptophan is present around the bacteria, it binds to the repressor and inhibits the entire process of transcription (section A of image 2).
The tryptophan which functions as a corepressor “switches off” the gene expression.
Contrary, when a bacterial cell lacks tryptophan, the corepressor can’t bind to the repressor. Hence, the repressor can’t bind to the activator for shutting down gene expression (section B of image 2).
This results in a “switch on” gene expression.
Conclusively, we can say the tryptophan itself regulates the mechanism of the repressible operon. Also, it is known as a negative type of operon.
The operon itself senses the presence or absence of tryptophan in a cell to avoid unnecessary gene expression.
It’s a natural mechanism, nature has the power to govern every cell’s metabolic and synthesis processes so errors may happen sometimes.
It might be possible that the corepressor isn’t able to activate the repressor to “switch off” the transcription. But don’t worry about that, the bacterial cell backed with another type of mechanism called attenuation.
If the entire mechanism of gene expression fails and RNA polymerase still synthesises the mRNA, the presence of high levels of tryptophan causes premature transcription.
Which means the RNA polymerase left the mRNA synthesis before it’s end. Or we can say it aborts the transcription before it reaches the stop codon. Therefore the gene for trp can’t form properly. This controls the tryptophan level in a cell.
The entire mechanism is known as attenuation. Furthermore, the trp operon is called a repressible negative regulation of gene expression.
lac operon:
The lac operon is the first operon well studied by scientists. F Jacob and J Monod won the Nobel Prize in physiology during 1965 for studying lac operon
The mechanism of the lac open is entirely different, we can say, entirely reverse than the trp operon. It is a kind of positive operon system.
Unlike the trp operon, here, gene transcription is turned “on” in the presence of the lactose- the corepressor.
Like glucose, lactose is yet another good energy source for bacteria, however, it uses lactose only in the absence of glucose. Here, in the lac operon, the lactose functions as a substrate for enzymes to promote transcription.
The promoter region is an RNA polymerase binding site, while the operator region is a repressor or inducer binding site.
The operon transcribed into polycistronic mRNA. Structurally, with a promoter, it has three different genes lacZ, lacY and lacA. All transcribed into single mRNA.
Along with these three genes, the lac operon also contains regulatory sequences that help in the transcription of genes.
Structural genes (LacZ, Y and A), promoter, terminator, regulator and operator sequences are key elements of the lac operon. See image 3 below,
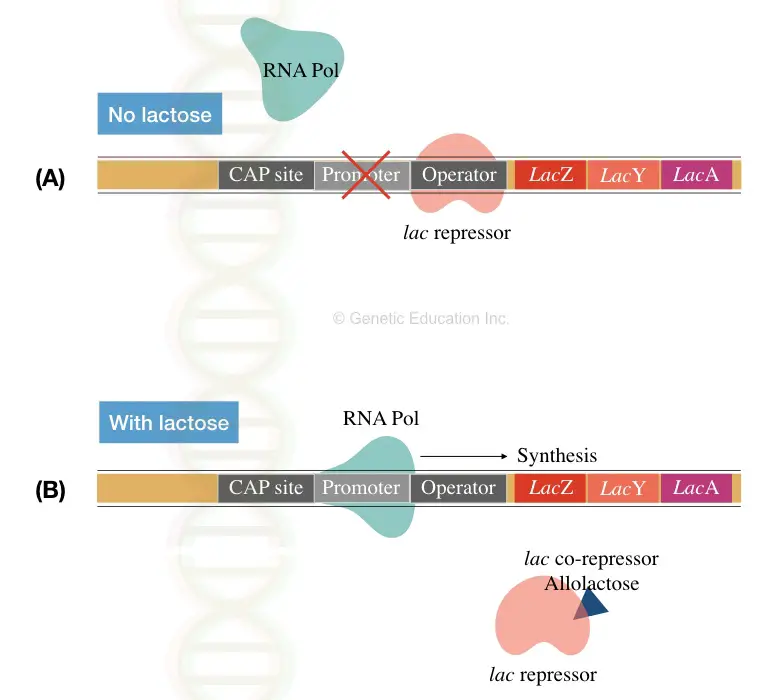
LacZ gene: Encode beta-galactosidase protein which cleaves lactose into glucose and galactose.
LacY gene: encodes beta-galactosidase permease protein- the membrane protein which helps in cellular transport of lactose into the cell.
LacA gene: encodes protein- galactoside acetyltransferase.
LacL: encodes lactose repressor protein
The lac operon is a type of negative inducible operon.
Here in the lac operon the beta-galactosidase encoded by the lacZ gene is required to convert the lactose into glucose and galactosidase- the disaccharides.
As we said, that the lac operon is used for metabolism and transportation of lactose into bacterial cells, another gene of this cluster, the lacY encodes the permease protein, which governs the cellular transport of lactose.
The lac operon is activated if only it is needed. Unlike the trp operon, it isn’t essentially required for a bacteria. It is activated “only” when lactose is available or glucose is not sufficient.
The whole mechanism functions by sensing the presence or absence of glucose or lactose.
In addition to this, the CAP- catabolite activator protein functions to sense glucose while the lactose repressor works as a sensory element for lactose.
The CAP binds the CAP binding site. It facilitates binding of RNA polymerase to the promoter region.
The CAP is also known as cAMP receptor protein. As the cAMP binds to the CAP, it induces the linking of CAP to the operon, in the absence of glucose.
Here when the repressor tightly binds with the operator in the absence of the lactose, the transcription is inhibited (see section A of image 3).
Contrary, in the presence of the lactose, the repressor detached from the operator sequence and allows RNA polymerase to transcribe the mRNA.
Here, the synthesis process is only possible because of the corepressor known as allolactose.
As we know, the corepressor changes the conformation of the repressor to control its function, the allolactose changes the shape of the repressor protein.
It can not bind to an activator, resultantly, RNA polymerase induces gene transcription (see section B of image 3).
Because of this reason, it is also known as inducible operon too. In normal condition, it remains “turned off”. But if lactose is present or glucose is absent, it “turns on”.
So let’s do not make the entire process more complicated. The point of all discussion is that “the lac operon is only activated when lactose is present in a cell or surrounding environment.” See these cases.
In the absence of glucose and the presence of lactose, Higher levels of transcription occur.
In the presence of glucose and the absence of lactose, no transcription occurs. Because lactose is absent, allolactose- corepressor of the lac operon can’t form. Therefore the entire transcription machinery shut down.
In the presence of glucose and lactose, less amount of transcription occurs. Depending upon the amount of lactose, the allolactose formed and regulated transcription.
In the absence of glucose and lactose both, no transcription happens.
Read more: DNA story: The structure and function of DNA
Conclusion:
Gene regulation process is essential for both prokaryotes and eukaryotes. Since the mechanism is different, the aim of it is to make a cell survive. Uncontrolled gene expression can cause various problems. It may cause cancer in eukaryotes.
Source:
Osbourn AE, Field B. Operons. Cell Mol Life Sci. 2009;66(23):3755–3775.