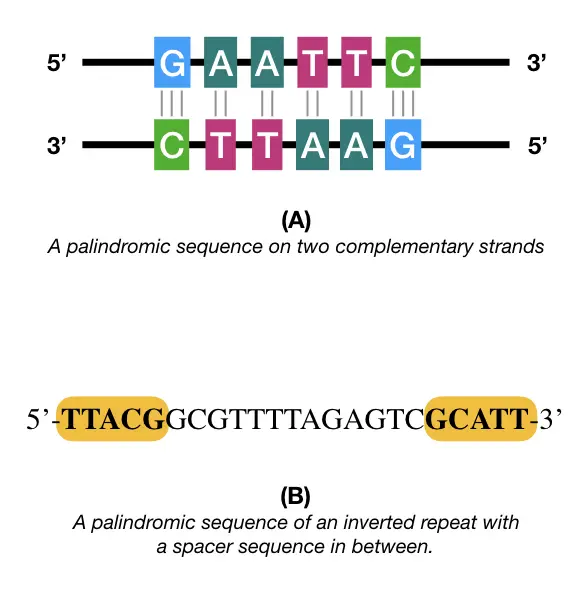
“A sequence when read from 5’ to 3’ and 3’ to 5’ direction on two antiparallel strands shows sequence similarity is said as the palindromic DNA sequence.”
Our genome, in fact, any genome on earth- made up of nucleic acid sequences; is amazing and mysterious. In layman’s terms, it is considered as a memory storing unit made up of only four nucleotides- A, T, G and C. We all know what these letters are!
The combination of these nucleotides forms various proteins and participates in regulatory processes such as transcription, translation and post-translational modifications.
In coding as well as in non-coding regions distinct patterns of sequence structures are observed. For example, the triplet codon CAG, repetitive nature of several sequences and dinucleotide repetitive units, and works as marker sequences to perform various investigations.
The palindromic nature of nucleotide sequence is one such pattern that is also a unique site for several enzymes to work and helpful for genetic studies. Put simply, when we read those sequences from one end to another and vice versa, it remains similar.
For example, 5’- GAATTC -3’, one palindromic sequence whose complementary sequence is 3’-CTTAAG-5’. Interestingly, the present sequence is also a restriction site for the EcoR1 restriction enzyme.
So what exactly the palindromic sequence is, how it is useful and what is its significance in our genome? all these questions and many more, we will answer in the present article. We will make sure that this article will have all the necessary information on this topic so that you do not have to search for more.
Stay tuned.
Key Topics:
What is a palindromic sequence?
Palindrome = similar from both directions
Or
Palindrome = reads the same from both sides.
As per the Oxford language dictionary, the “Palindrome” means
“A word or sentence read the same from backward and forward, for example, Madam, kayak or refer.”
Oxford language dictionary.
The same mentions have been applied to some of the genomic elements, a short one, usually non-coding and located one after another or randomly reads similar, as aforementioned in the example.
“I did, did I?”
“Eva, can I see bees in a cave?”
In biological science either in DNA, RNA or protein sequence-specific palindromic patterns are observed. Specifically, when considering the DNA, the palindromic sequences are either an exact match or partial. Take a look at the examples of each.
5’-GAATTC-3’
3’-CTTAAG-5’
5’-TTACGGCGTTTTAGAGTCGCATT-3’
The first example is a true palindromic while the other one is a type of an inverted repeat, often present in the transposable elements like LINEs and SINEs consisting of spacer sequences in between the two types of similar sequences.
In the human genome, the Y chromosome consists of a large number of palindromic repeats and as per the studies are also involved in several diseases (Anjana et al., 2013).
These sequences are often known as “self-complementary sequences” as well.
Definition:
A type of nucleotide sequence shows similarity when read from its complementary sequence in reverse order.
Or
A sequence reads similarly from 5’ to 3’ and 3’ to 5’ direction on both complementary strands.
Importance of Palindromic sequences:
Several important functions such sequences perform in our genome are,
- Gene expression and regulation
- Gene replication
- Homologous recombination
- The recognition site for restriction enzymes
- Binding sites for many enzymes and transcriptional factors.
- Stop transcription and initiate translation.
- It is prevalent in regulatory regions.
Significance of Palindromic sequences:
Studies suggest that the length of a palindromic sequence has a serious role in various genetic activities and is also involved in deletions, insertions or diseases.
Ganapathiraju et al., 2020 Explained in their paper entitled “A reference catalog of DNA palindromes in the human genome and their variations in 1000 genomes” that 30% of such sequences are prone to variations and are involved in single and multi-trait disorders.
In addition, their paper also suggests that palindromes lower than 50bp prevent genomic DNA degradation, on the other side, longer sequences than 50bp create genome instability thereby a disease.
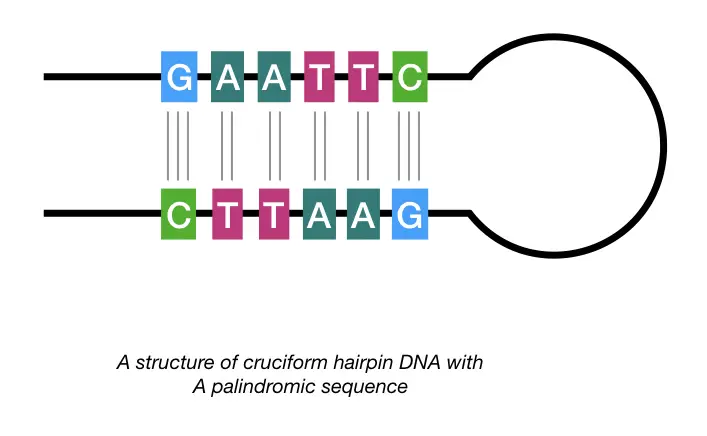
Henceforth, longer palindrome or near-palindrome sequences are not so suitable for our genome as they can form a hairpin. They form a hairpin-like secondary structure by folding back and forming a structure known as “cruciform”.
Studies evidence that cruciform hairpins are regions more prone to DNA breakage, deletion and translocations which are inherited as well. And results in disease conditions.
Near-palindromic sequences which are not exactly similar, as aforementioned, like the inverted repeats, commonly have cruciform hairpins due to having larger spacer elements within the palindromic elements. Take a look at the example,
Most of the palindromic sequences are actually a recognition site for so many restriction enzymes therefore commonly found in the prokaryotic genome but remained silent by methylation. Meaning, a bacterial REase can’t cut its own.
A recognition site is a kind of marker for the restriction enzyme to cleave the DNA, which is unique and identifiable. Here are some of the examples of various restriction enzymes and their recognition sites.
Examples of palindromic sequences:
| Restriction enzyme | recognition sequence |
| EcoR1 | GAATTCCTTAAG |
| BamH1 | GGATCCCCTAGG |
| Taq1 | TCGAAGCT |
| Alu | AGCTTCGA |
| HaeIII | GGCCCCGG |
| HindIII | AAGCTTTTCGAA |
| PstI | CTGCAGGACGTC |
| NotI | GCGGCCGCCGCCGGCG |
Note that each enzyme upon cutting produces either sticky ends or blunt ends, which is a separate topic for discussion.
Wrapping up:
Less published data are available on the importance of palindromic sequences, in fact, I didn’t find the name of the original scientists who discovered it. However, research studies depict a potential role in the disease or disease susceptibility.
Palindromic repeat sequences might be formed accidentally but have crucial importance for prokaryotes like bacteria as they possess some recognition sites and methylation sites to cleave or not to cleave the DNA.
Sources:
Anjana, R., Shankar, M., Vaishnavi, M. K., & Sekar, K. (2013). A method to find palindromes in nucleic acid sequences. Bioinformation, 9(5), 255–258.
Ganapathiraju, M.K., Subramanian, S., Chaparala, S. et al. A reference catalog of DNA palindromes in the human genome and their variations in 1000 Genomes. Hum Genome Var 7, 40 (2020).


