“Explore the concept of DNA methylation. Learn the basics, importance and detection techniques for DNA methylation in this article.”
DNA methylation is an epigenetic modification. Epigenetics is defined as any changes in our DNA that impact gene expression and regulation. Several common epigenetic modifications are histone modification, chromatin remodeling, DNA packaging, etc.
Epigenetic modifications are different from genetic alterations as those aren’t structural changes. Meaning, it can not alter the structure of a gene, rather, it alters how it is expressed.
When a gene produces a protein, it is considered “expressed.” Epigenetic modifications directly impact protein synthesis. Methylation, a prevalent epigenetic modification found throughout our genome, is one example of these modifications.
Research suggests that 1% of our genome contains methylated DNA sequences. So, what exactly DNA methylation is, how does it occur and how important is it?
Let’s find out.
Related article: Epigenetics 101: What is Epigenetics and How Does It Work?
In this article, we’ll explain the concept of DNA methylation from scratch. This article is for beginners but they will also learn all important information related to the topic.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is DNA Methylation?
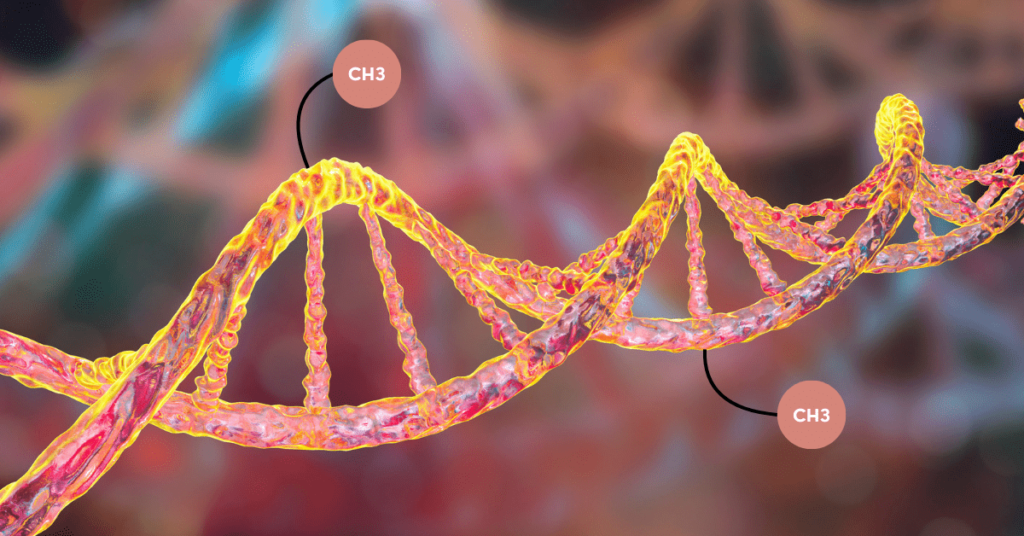
Methylation is a chemical process in which a methyl group (-CH3) is incorporated in the reaction or on a molecule. As every biological reaction is catalyzed by an enzyme, methyltransferase governs the present incorporation process.
Now, coming to our DNA.
Our DNA is a long polynucleotide chain, made up of nucleotides. Each nucleotide has a basic structural unit- a nitrogenous base (+ sugar + phosphate). This base is a ring structure and when a methyl group (-CH3) is added to it, it’s referred to as DNA methylation.
In particular, the methyl group is incorporated into any of the nitrogenous bases. Adenine and Cytosine can be methylated in mammals. However, cytosine is the most common target for DNA methylation.
Interestingly, DNA methylation was discovered a few years before the actual discovery of DNA and its chemical structure. In 1948, Rollin Hotchkiss identified modified methylated cytosine during their research.
Their research initially suggested that this modified base, 5-methylcytosine is naturally occurring in the DNA. However, its role in gene expression and regulation was unknown. Later on in 1980, studies came into the picture that depicted the role of DNA methylation in gene expression and regulation.
DNA methylation results in the downregulation of gene expression, while DNA demethylation leads to the upregulation of gene expression. Recent research has highlighted that dysregulation of the DNA methylation mechanism contributes to various types of cancer.
How Does DNA Methylation Occur?
DNA methylation is a fundamental biological process. As aforementioned, the addition of a methyl group to the cytosine base of DNA leads to DNA methylation.
A group of enzymes known as DNMTs (DNA methyltransferases) catalyze the process. Depending on, at which stage the DNA methylation occurs, a different DNMT will take part in the process.
We will discuss various types of DNMTs in the next sub-section. We’ll first understand the molecular mechanism.
Now, for the present event to occur, what is the primary requirement? A methyl group, Right? So how and from where it is sourced?
The methyl group for various methylation events including DNA methylation has been sourced by SAM (S-adenosylmethionine). SAM is synthesized in a cell through various biochemical pathways.
Next, how can a chemical group be incorporated? using the help of an enzyme. The enzyme DNMT (DNA methyltransferase) catalyzes the present biochemical reaction. DNMT first, finds out the target location, recognizes the cytosine and initiates a catalytic reaction.
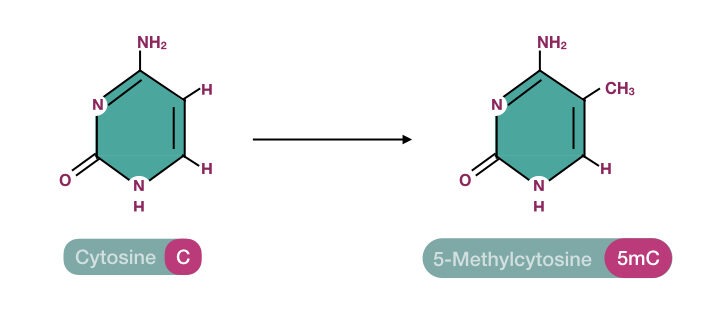
Here, the SAM binds to the enzyme’s active site. Now the enzyme performs chemical modification and transfers the methyl group from SAM to the 5th carbon of the cytosine. Here the CH3 group is covalently attached to the 5th carbon of the cytosine ring and forms 5-methylcytosine.
The usual target site for methylation is CpG-rich genomic regions which are the main body of gene promoters and other regulatory elements. Once again, you may have a question, what is the 5mC and its significance?
The fifth base:
~1.5% of our genome has methylated cytosine bases (5mC), therefore, is considered as a 5th DNA base. However, it’s just a chemical modification of a base. The natural replication process can not incorporate it in a DNA sequence and hence can not be considered as a true or natural DNA base.
Still, 5mC has a significant importance in gene expression and regulation. 80 to 85% of the mammalian CpG islands contain 5mC. The presence of the methyl group prevents enzymes from initiating gene expression activities.
Research suggests that 5mC is abundantly present in a DNA major groove and prevents enzymatic bindings for gene expression. Noteworthy, in bacteria, 5mC also has an additional function. It protects the bacterial DNA from restriction digestion by its own restriction enzyme.
DNA Methylation Enzymes
DNA methyltransferases (DNMTs) are the enzymes used in various DNA methylation activities. DNMTs are important for maintaining and producing DNA methylation patterns. It can also make new DNA methylations based on the requirements.
Deletion studies in mice showed that either one or all DNMT deletions result in mice’s fetal death in the early embryonic stage. This signifies the importance of DNMT enzymes for an organism.
DNMTs play a vital role in genome stability and integrity. Inadequate activities of any DNMTs result in chromosomal instability and cancer. They methylate genomic regions including CpGs, transposons, imprinted genes, the X-chromosome and other non-coding regions.
Four different DNMTs in mammals are DNMT1, DNMT3A, DNMT3B and DNMT3L.
DNMT1:
DNA methyltransferase 1, present in mammals, is also known as a maintenance enzyme. Let me tell you why! It perverses and inherits the original methylation pattern.
DNMT1 works during the process of replication, where the DNA is copied and distributed into daughter cells. During replication, the hemimethylated DNA is copied. DNMT1 is situated near the replication fork and methylates the hemimethylated newly synthesized DNA.
This means it prepares the same methylation pattern, as the original DNA sequence has. In addition, it also helps in DNA repair activities as well.
DNMT1 is expressed at a higher rate during the S cell division phase and in various proliferating cell types. For instance, embryonic, hematopoietic and epithelium stem cells. Recent findings also suggest that DNMT1 is found in brain tissues in very high amounts.
DNMT3
Two common types of DNMTs- DNMT3A and DNMT3B are also present in the mammalian methylation process. Both present methylation enzymes are considered as de novo DNA methyltransferases.
DNMT3A and DNMT3B can methylate the already existing as well as new DNA sequences. Meaning, they can produce new DNA methylation patterns, new genotypic combinations and resultantly, diverse phenotypes.
Despite the extreme similarity in structure and function, both are expressed differently in different tissues. Moreover, the DNMT3A is also expressed during embryonic development but during later stages.
DNMT3A further participates in chromatin remodeling as well as in the demethylation process.
DNMT3B on the other hand, is expressed during the early developmental stage. Its higher expression is reported in germ cells, bone marrow and pluripotent stem cells, in particular.
Mutations in DNMT3B lead to immunodeficiency, centromeric instability and facial anomalies syndrome.
DNMT3L
DNMT3L lacks the catalytic domain thereby the catalytic activity. However, it elevates the activity of DNMT3A and DNMT3B during the developmental stage. A higher expression of DNMT3L is reported during the developmental stage.
Studies also showed that the association of DNMT3L with DNMT3A and 3B is important for maternal and paternal imprinting.
Function of DNA methylation:
DNA methylation is an important epigenetic alteration and crucial for our genome. It is extensively involved in gene expression and regulation. To do so, DNA methylation is significantly reported on CpG, intergenic and gene body parts. It’s also reported during X chromosome inactivation.
We will prepare a detailed and separate article explaining the role of DNA methylation.
| DNMT Enzyme | Function |
| DNMT1 | Maintenance methyltransferase |
| Recognizes hemi-methylated DNA strands during DNA replication | |
| Adds methyl groups to newly synthesized DNA strands to maintain methylation patterns | |
| DNMT3A | De novo methyltransferase |
| Responsible for establishing new DNA methylation patterns during embryonic development and cellular differentiation | |
| DNMT3B | De novo methyltransferase |
| Works in conjunction with DNMT3A to establish DNA methylation patterns | |
| Plays a role in genomic imprinting and X-chromosome inactivation | |
| DNMT2 | tRNA methyltransferase |
| Catalyzes the methylation of cytosine residues in tRNA molecules | |
| Not directly involved in DNA methylation processes | |
| DNMT3L | Helps DNMT3A and DNMT3B during the catalytic reaction. |
| Lacks catalytic domain. |
Detection Techniques
The present DNA modification plays an important role for us. The dysregulated methylation process causes cancer and other problems. Various techniques that scientists use for quantitative and qualitative analysis of DNA methylation are; Methylation-specific PCR and Methylation sequencing.
Methylation-Specific PCR:
Methylation-specific PCR is a specialized technique, developed for DNA methylation analysis. Followed by bisulfite treatment, the technique studies a gene or genomic region containing DNA methylation.
A simple PCR-based analysis can be conducted by preparing bisulfite-converted primers and normal primers. To know more about the bisulfite conversion, click the link and read the reaction.
This will help to determine, if methylation occurred or not. When this whole experiment is performed on a quantitative PCR, the difference between methylated and non-methylated amplification ct value will help estimate relative DNA methylation.
High-resolution melting curve analysis is yet another PCR analysis that helps to estimate DNA methylation. SYBR green dye is usually used during probe hybridization to the target region, but for analysis of the template dissociation, a high-resolution melting curve is prepared for.
The present method is an easy, quick and reliable quantitative DNA methylation analysis method, but at the same time, can only perform analysis for a few CpGs or gene promoters.
Some literature also suggests one PCR modification known as COLD-PCR (Co-Amplification at Lower Denaturation Temperature). It allows the amplification and detection of minor or rare alleles present in the sample.
It can detect rare and low-level unmethylated DNA from the methylated DNA. Here, the low-level unmethylated DNA is amplified efficiently at a lower denaturation temperature. Due to this reason, it is highly sensitive and is used in diagnostics.
Bisulfite sequencing:
Bisulfite sequencing is another powerful approach for studying DNA methylation. It can provide not only quantitative data but also sequence-specific DNA methylation information.
Again the process is started with bisulfite sequencing. The bisulfite-converted DNA sequence is compared with the normal or reference sequence. Such analysis helps in the methylation quantification, differential methylation and methylation-specific sequence alterations study.
Targeted bisulfite sequencing is used to study DNA methylation from a specific gene or sequence while whole-genome bisulfite sequencing is used to study DNA methylation from the entire genome. To learn more, read this article: What is Bisulfite Sequencing?- Beginners To Advance Guide.
Restriction Digestion-Based Method:
Another popular approach is using the restriction enzyme to detect the methylation of DNA. some particular restriction endonucleases, for instance, MspI can cleave the CCGG-rich regions of the genome.
These sequences are CpG-rich, when combined with the sequencing, the present technique can efficiently detect the methylation in CpG-rich DNA. The present method is known as Reduced Representation of Bisulfite Sequencing (RRBS).
Click the link to read this topic in the bisulfite sequencing article.
Wrapping up:
DNA methylation is an important epigenetic modification that regulates gene expression, however, the dysregulation can cause serious health-related problems, particularly, cancer.
Differential methylation pattern is observed in different tissues and even in different cells. So it can perform tissue-specific gene regulation. Notwithstanding, more studies are required to explore more about the present modification, and develop new assays and testing methods.
I hope you like this article. Share this article and follow the article links provided in this article to strengthen your knowledge of epigenetics and DNA methylation.
Sources:
Moore, L., Le, T. & Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacol 38, 23–38 (2013). https://doi.org/10.1038/npp.2012.112.
Chen Z, Zhang Y. Role of Mammalian DNA Methyltransferases in Development. Annu Rev Biochem. 2020 Jun 20;89:135-158. doi: 10.1146/annurev-biochem-103019-102815. Epub 2019 Dec 9. PMID: 31815535.
Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3-29. doi: 10.1007/978-1-4419-9967-2_1. PMID: 22956494; PMCID: PMC3707278.
Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel). 2016 Jan 6;5(1):3. doi: 10.3390/biology5010003. PMID: 26751487; PMCID: PMC4810160.