“Methylation-Specific PCR is employed to study DNA methylation. Learn the concept, experimental setup, types, advantages and limitations of Methylation-Specific PCR.”
An epigenetic modification, DNA methylation regulates gene expression. DNA methylation analysis and detection help scientists understand the expression of a gene or genome.
DNA methylation controls early development, imprinted gene expression, X-chromosome inactivation and many other biological activities. Restriction digestion, PCR and Bisulfite sequencing are several techniques used to study it.
Among various available techniques, methylation-specific PCR is one popular technique that has been utilized to study DNA methylation. It’s a bisulfite conversion-based technique that can amplify and quantify the CpG-rich methylated regions, effectively.
In this article, I will explain to you the concept of methylation-specific PCR, its principle, process, primer design, and variations. I will also discuss the advantages and limitations of MS-PCR in this article.
This article is for students who are curious about MS-PCR as well as for researchers and budding scientists who want to utilize the present technique in their research projects.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is MS-PCR?
Methylation-specific PCR is the most common and preliminary method used to study a particular gene’s promoter methylation status. It enables methylation analysis for a single or a few genes. We use MSP in short for methylation-specific PCR, throughout the present article.
MS-PCR is Methylation-Specific PCR and is used to study DNA methylation. In 1996, Herman et al. explained the methylation-specific PCR technique for amplification of CpG-rich genomic regions.
However, TaqMan probe-based qRT-PCR was performed by Eads et al (1999). It’s also noted that the actual chemical modification, DNA methylation was identified before the discovery of DNA.
Conceptually, the MSP looks like a simple assay but it is a bit complex process. Unlike the normal PCR, here two separate PCR primers have been used for two separate PCR reactions.
One set of primers is specific to the bisulfite-treated DNA while another set of a primer is specific to the non-bisulfite-treated/methylated DNA. Differential banding pattern helps understand the methylation status of the target sequence.
We will understand the results, in the results and analysis part, separately.
A few steps in the present technique are DNA extraction, primer design, bisulfite conversion, PCR reaction preparation, PCR amplification and post-PCR analysis. Bisulfite conversion for DNA methylation was already discussed in our previous article. You can read it here from the link.
Principle of MS-PCR
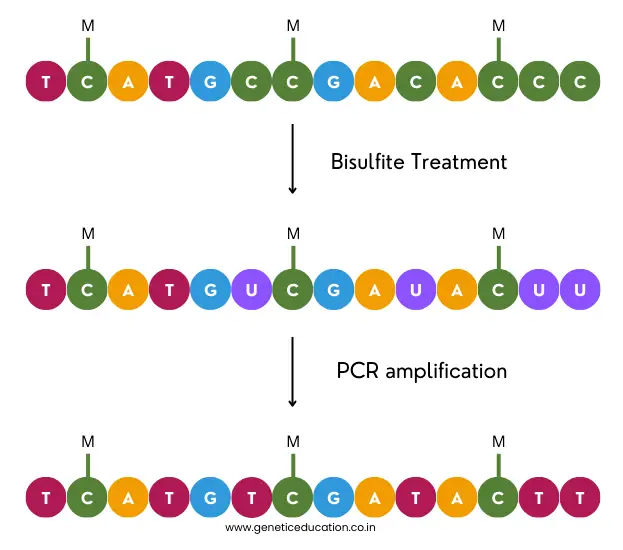
Methylation-specific PCR is capable enough to detect the methylation pattern associated with a specific gene. In principle, the bisulfite treatment converts the non-methylated cytosine into uracil which is detected by the non-methylated DNA-specific primer set.
Contrary, the methylated DNA remains untreated with the bisulfite and can be detected by the methylated DNA-specific primer set. Using a conventional agarose gel electrophoresis assay, or qPCR assay, the methylation status can be determined.
Process and Steps:
Steps in the complete MSP process are DNA extraction, bisulfite conversion, primer design for MS-PCR, PCR amplification and detection. Individual steps are explained here.
DNA isolation
MSP requires a higher amount of genomic DNA, usually 100nG to 2µG. Although a well-standardized DNA extraction protocol can yield a sufficient amount of genomic DNA for diagnostic experiments, I strongly recommend using ready-to-use DNA extraction kits.
Spin-column DNA extraction kits can yield a good quality and quantity of genomic DNA. Furthermore, it doesn’t require any post-purification step.
Bisulfite conversion
In the next step, the DNA is treated with the bisulfite reagent. It will convert the non-methylated cytosines from the CpG-rich region to uracil while leaving the methylated DNA untreated.
The whole process and protocol are discussed in our previous article. You can read it here as well: Bisulfite treatment for DNA methylation.
With a well-optimized and standardized protocol, the present assay can be used in the diagnosis. One factor among many that is considered for standardization is primer design.
Primer Design for MSP:
The first step in the MSP primer design process is identifying the promoter region for a gene of interest. Many databases contain such information. A promoter sequence can be extracted from any of these databases and used for MSP.
Several rules for MSP primer design are listed here.
- Target the CpG island of a gene promoter region.
- CpGs are highly susceptible to DNA methylation. Primers should flank the CpG island of a gene but avoid the boundaries.
- The CpG boundaries can impact the amplification efficiency so avoid it.
- The primers should be located in the vicinity of the Transcriptional Start Site (TSS). Research suggests that the 1000 bp upstream and 500 bp downstream regions of TSS are good choices for primer design.
- The primer should effectively discriminate between methylated and non-methylated sequences.
- MS-PCR primers should be longer than the normal PCR primers. Particularly, the non-methylated primer which contains the AT-rich counterpart. The methylated primer should be shorter while the non-methylated primer should be longer.
- The ideal length of the non-methylated primer should be between 20 to 30 nucleotides.
- Each primer should contain at least a single CpG-rich complementary region at the 3’ end, however, both primers should have the same number of CpGs.
- The primer sets are designed in such a way that the PCR products should not exceed more than 300bp.
- The melting temperature between both the primer sets should not exceed more than 5ºC.
- It’s also important to note that the primer should also contain an adequate number of non-CpG ‘C’ nucleotides to effectively amplify the bisulfite-converted DNA.
Other rules associated with the common PCR primer design are also applicable in this case. MethPrimer and Bisulfite Primer Seeker by ZymoResearch are two amazing and free tools available online for MSP primer design.
PCR amplification:
Usually, the PCR reaction preparation and amplification steps in MSP will remain the same as in other normal PCR experiments. All the important ingredients including dNTPs, PCR buffer, primers, Taq DNA polymerase, DNA and water are added.
The amplification conditions are set as per the protocol provided by the MSP kit provider or you can simply use a general protocol and temperature conditions. However, keep in mind that the annealing temperature is set as provided by the primer design tool.
MS-PCR detection:
The methylation-specific PCR reaction can be analyzed using a 2% agarose gel. The methylated and non-methylated reactions can be run on a gel side-by-side and analyzed under a UV transilluminator.
Notedly, quantitative PCR uses melting curve analysis for quantifying the number of methylated and non-methylated templates present in the sample.
How to Read Methylation-Specific PCR Results?
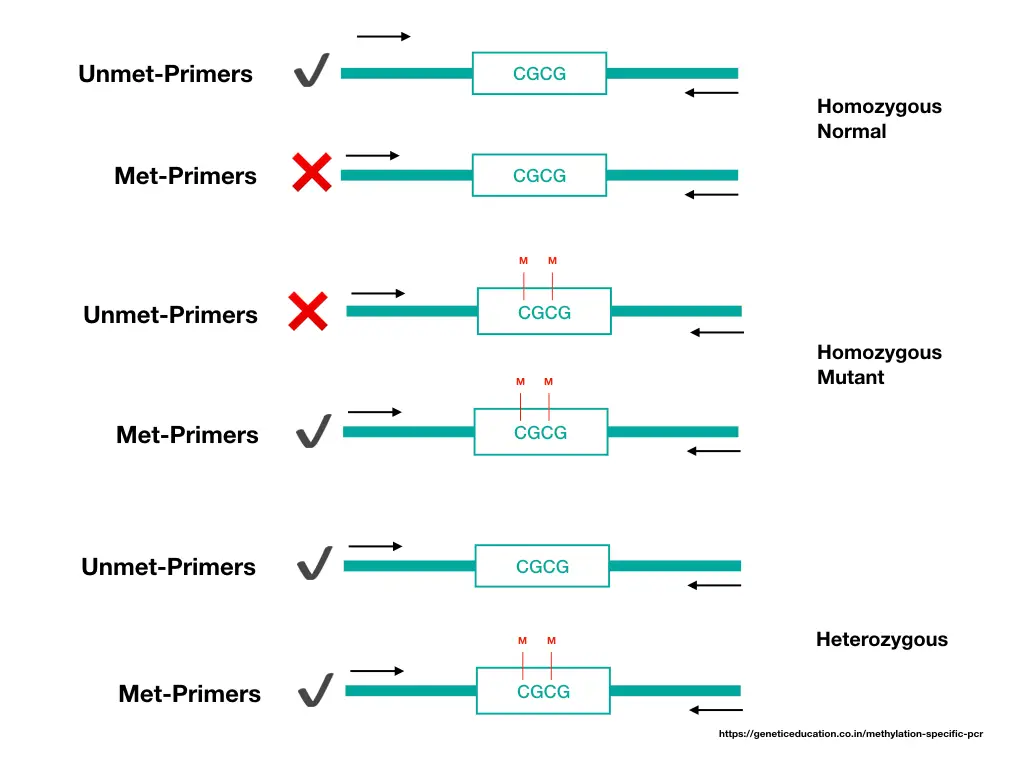
Now let’s take one hypothetical example to understand the MS-PCR results. Check out this image first,
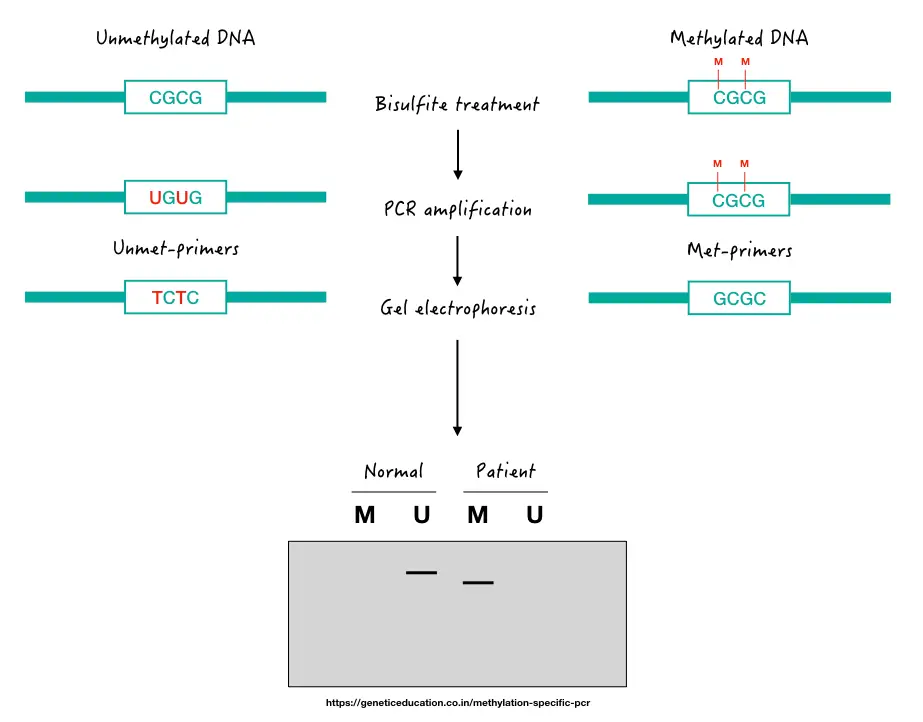
Here, as aforementioned, we use two sets of primers, one specific to non-methylated (normal DNA) and another specific to methylated DNA (patient DNA). If methylation occurs on the target gene promoter, it will give us a band in the ‘M’ lane of the patient’s sample.
If not, it will give us a DNA band in the ‘U’ lane of the gel, just like the normal sample. For instance, suppose, we want to know the methylation status of the P53 gene promoter for one type of leukemia.
Now, if methylation occurs in the patient sample, it will give us a DNA band in the ‘M’ lane, otherwise in the ‘U’ lane. Notedly, DNA band size will depend on the number of CpGs present in the promoter.
In the case of a heterozygous condition, in which one allele is methylated and another lacks methylation, we can get two DNA bands in ‘U’ and ‘M’ lanes.
MS-PCR Variations:
What we have discussed so far is just a conventional approach to studying DNA methylation for a gene. Many variations of the native MSP are now available depending on the requirement of the experiment.
Here I am explaining some of the popular, important and well-validated MSP variations.
Nested MSP:
Nested PCR is utilized to improve the detection sensitivity with precision. Here, it enables the detection of low-level DNA methylations. The experimental scheme will remain the same as the conventional nested approach.
With the two sets of primers (unmet and met-primers) another set of primers as outer primers is used. The outer primers will amplify the larger flanking region of the target CpG region.
This PCR product serves as a template for our MSP primers that will amplify their respective allele with their respective set of primers. The present optimization can improve the overall sensitivity of the experiment. To know more, you can read this article: What is Nested-PCR?- Concept, Primers, Protocol, Advantages and Limitations.
Notedly, using many primers in a single reaction can complicate the reaction preparation.
Multiplex MSP:
The limitation of the conventional MSP is the throughput. Only a single or few methylation sites can be analyzed. Using a multiplex MSP approach methylation patterns within various genomic regions can be investigated in a single reaction.
To do so, for each site, a dedicated primer set is prepared. MMSP can efficiently determine methylation status from various CpG-rich regions, methylation from various genes or dense CpG-rich regions.
Put simply, researchers can study multiple methylation sites in a single run by preparing a single reaction. However, multiplex PCR can result in false positive and false negative results.
Utilizing many primer sets in a single reaction can complicate the reaction preparation and analysis procedure. Expertise is needed to design and perform such experiments.
Quantitative MPS:
Conventional MPS is unable to provide quantitative data. Meaning, that we can only determine if methylation is present or not but can not determine the quantitative. qMPS- quantitative methylation-specific PCR is a technique that can quantify the amount of methylation.
Two common assays used for real-time methylation monitoring and quantification are the dye-based method and the probe-based method. The hybridization probe-based method is more effective than the intercalating dye-based approach.
qMSP allows real-time quantification of methylated and non-methylated templates present in the sample. Put simply, it measures the methylation amount at a particular CpG locus.
Such quantitative data can be compared with the reference data or across other samples to understand the condition associated with the methylated locus. In addition, qMSP data are also used to correlate the amount of methylation with the associated disease severity.
The present quantitative method is highly sensitive, accurate, time-saving and cost-effective. qMPS is used in diagnostics.
MS-HRM:
Conventional MSP requires post-PCR analysis, for instance, gel electrophoresis. MS-HRM (Methylation-specific high-resolution melting curve analysis) is an RT-PCR-based method that doesn’t require any post-processing.
Methylated and non-methylated PCR amplicons are simultaneously detected by their template melting properties. In the present intercalating dye-based approach, methylated and non-methylated amplicons are detected using their melting temperature differences.
The amplified templates are melted based on their melting temperature, each template is quantified. The present approach is suitable for high-throughput DNA methylation analysis.
COBRA:
Combined Bisulfite Restriction Analysis (COBRA) is a qualitative DNA methylation analysis technique that requires restriction digestion. As this procedure includes both PCR amplification and restriction digestion, it’s considered RFLP for DNA methylation.
Here, the bisulfite-treated DNA is amplified in the PCR reaction using the same conventional PCR approach. Now, the amplicons are selectively digested using a restriction enzyme.
Restriction enzymes can not cut methylated DNAs thus, the REase was selected only to cut the non-methylated DNA. Meaning, that in a gel, various fragments, depending on the number of restriction sites, are obtained for non-methylated DNA.
Such qualitative assessment helps understand the presence and absence of DNA methylation. However, it can not be used in diagnosis, as it can not provide qualitative data only.
COBRA is a cost-effective and simple method. It can be used to investigate DNA methylation for a smaller number of CpG sites.
Summary:
| Types of MSP | Specialty | Advantages | Limitations |
| Conventional MSP | Detects methylation through amplification using two primer sets. | Simple and cost-effective. | Require separate PCR reactions, lack multiplexing, and lack quantitative analysis. |
| Nested PCR | Use a nested set of outer primers for precise amplification of target location. | High sensitivity and specificity. Can detect low abundant methylated templates.Suitable for samples with low DNA methylation levels. | Requires an additional set of primers, prone to contamination, increased risk of non-specific amplification. |
| Multiplex MSP | Use multiple primer sets. | Detect many CpG regions, and reduce starting material requirement and experimental variability.High throughput analysis. | Complex setup. Prone to primer-dimer, non-specific amplification and false results. |
| Quantitative MSP | Probe or dye-based quantification approach. | Provides quantitative data, high sensitivity and specificity and differential methylation analysis. | Requires fluorescence chemistry, costly and can not provide sequence-based data. |
| MS-HRM (High-Resolution Melting Curve Analysis) | Dye-based approach for melting template analysis. | post-PCR is not needed, high throughput, sensitive and accurate. | Can not provide sequence-based data. |
| COBRA | Used restriction digestion for analysis. | Cost-effective and easy method. | Limited throughput and analytic sensitivity. Can not provide quantitative data. |
Advantages of MSP:
- Now in this section, I will explain the advantages of MSP in general.
- Methylation-specific PCR is a sensitive assay that can detect low DNA methylation levels effectively.
- It’s a cost-effective and time-saving procedure. Extraction to analysis can take 4 to 5 hours only.
- It is highly specific as well, as different primer sets are designed for methylated and non-methylated regions as well as for different CpG regions.
- The present approach provides qualitative and quantitative DNA methylation data and hence is used in diagnostics as well.
- The amplicons can be used directly for bisulfite sequencing.
Disadvantages of MSP:
In this section, I will explain the disadvantages of MSP in general.
- Methylation-specific PCR has a limited throughput. It can analyze a small number of CpGs or methylated sites.
- Techniques like conventional or nested approaches needed additional post-processing like gel electrophoresis.
- The results can be influenced by many factors such as primer design, reaction preparation and PCR conditions.
- It has a higher chance of contamination and false results.
- It can not provide sequence-based methylation information.
Wrapping up:
MSP- Methylation-Specific PCR is a decent technique to utilize for small-scale DNA methylation studies. Although it can provide amazing qualitative and quantitative data, it can not give us more information.
Also, it has limited throughput and can not provide genome-wide data for methylation. Still, MSP is widely used in diagnostics for cancer studies and other methylation-specific analyses.
I hope this article added much value to your DNA methylation analysis. We complete this series here. You can read our previous articles on bisulfite conversion and bisulfite sequencing to get more knowledge on this topic.
Resources:
Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9821-6. doi: 10.1073/pnas.93.18.9821.
Wani K, Aldape KD. PCR Techniques in Characterizing DNA Methylation. Methods Mol Biol. 2016;1392:177-86. doi: 10.1007/978-1-4939-3360-0_16.
Ku JL, Jeon YK, Park JG. Methylation-specific PCR. Methods Mol Biol. 2011;791:23-32. doi: 10.1007/978-1-61779-316-5_3.
Yoshioka M, Matsutani T, Hara A, Hirono S, Hiwasa T, Takiguchi M, Iwadate Y. Real-time methylation-specific PCR for the evaluation of methylation status of MGMT gene in glioblastoma. Oncotarget. 2018 Jun 12;9(45):27728-27735. doi: 10.18632/oncotarget.25543. PMID: 29963232; PMCID: PMC6021237.