“Long-read sequencing allows precise analysis of complex genomic regions and alterations. Learn about the importance and applications of long-read sequencing in this article.”
‘Reads’ in sequencing is an important terminology that simply represents nucleotide sequencing. When sequencing is read, that means, the machine is sequencing the nucleotides present in the sample or fragment.
In a more simplified version, we can say, “The machine is reading the fragment or sequence.” Depending on the length of a fragment, going to be read by the machine, sequencing reads can be divided into short and long sequencing reads.
Different platforms have varied read lengths to achieve higher precision in sequencing. For example, Sanger sequencing, HiSeq, MiSeq and NovaSeq have shorter read lengths while PacBio’s SMRT and Oxford Nanopore sequencing have long read lengths.
Each short and long-read sequencing has its own advantages and limitations. However, in routine clinical applications, short-read sequencing is more popular. Still, long-read sequencing has pivotal importance as well.
In this article, I will comprehensively explain the concept of long-read sequencing, its advantages, limitations and applications.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is Long-Read Sequencing?
The third generation of sequencing, long-read sequencing is a chemistry in which larger DNA fragments of a few kilobase pairs are sequenced effectively. Ranging from 1000 bp to 10,000 bp or even 100,000 bp fragments can be sequenced.
Various chemistries have been developed for doing long-read sequencing. However, to understand the importance of long-read sequencing, we first need to know why long-read sequencing is required.
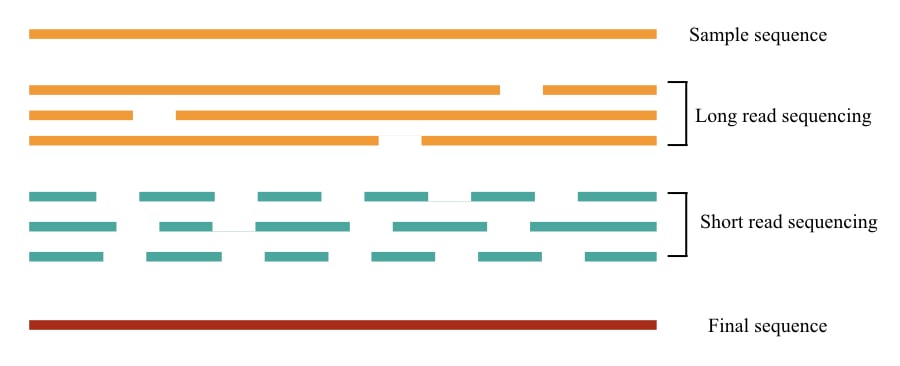
Why is Long-Read Sequencing needed?
Sequencing is a complex process. Still, it’s comparatively easy to sequence shorter DNA fragments. A sequence of 30 to 100 bp can be ‘nearly’ accurately sequenced. The reason is that it’s short; the machine has to invest less power to read and sequence the fragment.
However, when it comes to complex genomic regions such as high GC-rich regions, and repetitive sequences, short read sequencing faces challenges. On a technical level, when it comes to assembling, such fragments are difficult to arrange and assemble.
Consider a scenario where a 2.5 kilobase (Kb) segment of the genome exhibits high GC-rich repetitive patterns. In such instances, sequencing, reading, and assembling this segment pose significant challenges for the sequencer. The machine may introduce sequence bias, leading to uncertainties regarding the accuracy of the sequence read.
Now, imagine, what if the whole 2.5kb fragment can be sequenced in a single run? That’s pretty good, right? Because it eliminates the uncertainty of ambiguity in assembling.
Indeed, the emergence of long-read sequencing addresses such challenges. Scientists leverage long-read sequencing to study complex genomic regions characterized by inversions, translocations, and deletions, as well as repetitive and high GC-rich sequences.
Related article: Genomic DNA Library- Preparation and Applications.
Types of long-read sequencing
As aforementioned, the majority of sequencing platforms use short-read chemistry. Interestingly, PacBio’s SMRT sequencing and Oxford nanopore sequencing use long-read sequencing chemistry.
These two platforms are true long sequencing platforms, however, several other methods such as sequencing by synthesis (Illumina) long-read sequencing chemistry are different and can not be considered true long-read sequencing.
True long-read sequencing reads the whole few kb long DNA fragment in a single run, while in other platforms, either short fragments are read and assembled as a long fragment or read as a long fragment and assembled in short reads.
PacBio’s SMRT sequencing and Oxford nanopore sequencing both sequence, read and assemble the whole long DNA fragment making them better at studying inversions and translocation, repetitive regions and high GC-rich fragments.
Long Read Sequencing Chemistry
In this section, we will elaboratively discuss various chemistries used by various platforms for long-read sequencing.
PacBio’s SMRT long-read sequencing
PacBio’s SMRT (Single Molecule Real-time) sequencing is also known as HiFi sequencing and works on the nanofluidic design. In this chemistry, first, the longer DNA fragment is circularized using known sequence adaptor ligation.
The polymerase is attached to each circular fragment during the library preparation and applied to the SMRT cell or chip. Each circular DNA fragment + polymerase complex is settled on the bottom of wells known as ZMWs (Zero-Mode Waveguides).
Now, nucleotides incorporated by the polymerase emit light signals which are captured by the detector and converted into the nucleotide sequence. Each incorporated nucleotide is read in real-time and thus the name SMRT sequencing has been given.
As the fragment is circularized, the polymerase continuously sequences the fragment in a never-ending process.
Using circular consensus sequencing, a 99% highly accurate long read sequence has been generated. While using continuous long-read sequencing, highly accurate and longest reads (>50Kb) can be generated.
Oxford Nanopore sequencing:
Among all the sequencing chemistries available, nanopore sequencing is interesting and looks easy. Here, the enzyme unwinds the double-stranded DNA and allows the single-strand to pass through the pore present in the membrane.
Nanopores on a membrane are submerged in the salt solution. When salt passes through pores, it completes the circuit and establishes current. Interestingly, when single-stranded nucleotide bases- A, T, G and C pass through the pores, they block the current in different ways.
This disruption is read and converted into the sequence data. Nanopore sequencing is a feasible option for long-read sequencing.
Illumina long-read sequencing
Illumina’s long-read sequencing can not be considered a true long-read sequencing, but it also does the job. Here, long and larger DNA fragments are marked using the enzyme.
These marks are used as a landmark read during the assembling of the fragment. Finally, all the marked regions are aligned and generate a longer original DNA sequence.
Do you Know?
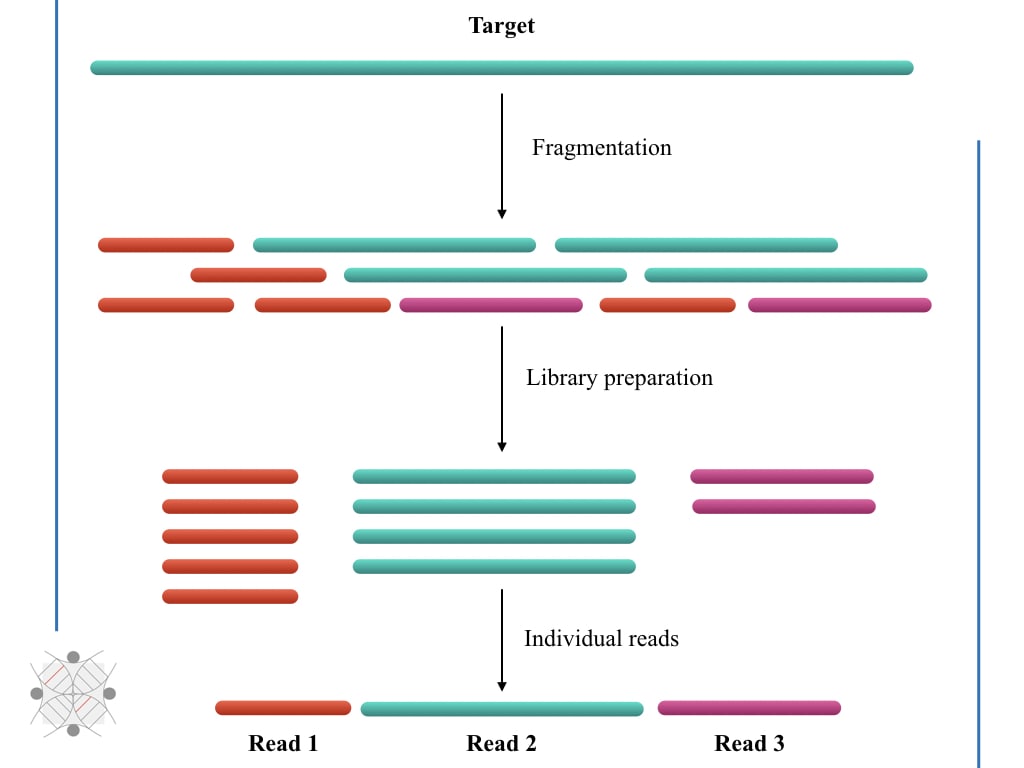
Advantages and Applications:
The present sequencing technique has been widely used in research but has less significant applications in clinical setups.
Genome-wide variant detection:
One amazing application of the present technique is to accurately investigate genome-wide variations such as larger deletions, duplications, translocations and inversions.
Studies suggest that long-read sequencing is an important tool for investigating structural alterations and complex rearrangements. Studies using PacBio technology have identified unique and large insertion and deletion events from the genome.
Sequencing of repetitive genomic regions:
Highly polymorphic and repetitive regions are hard to sequence and study. Long-read sequencing can effectively identify variations in repetitive, tandem and highly polymorphism regions.
Epigenetic studies:
Long-read sequencing like HiFi sequencing has a unique application in analyzing epigenetic alterations. These platforms don’t rely on any amplification step, so technically, the ‘base’ is sequenced as it is.
Any modification present in a base, for example, methylation can be tracked and studied using a PacBio HiFi long-read sequencing. The present advantage helps to study the heritable gene expression changes and other epigenetic alterations.
Haplotype phasing:
The present technique is also used for haplotype phasing to investigate the inheritance of maternal and paternal DNA sequences, alterations or sequence variations.
As per PacBio the accuracy of long read sequencing is 99.9%.
Read more: A Guide To Next-Generation Shotgun Sequencing In Metagenomics: Technique, Advantages and Challenges.
Limitations:
Challenges in data analysis:
Short read sequencing is popular, convenient and effective thus most platforms and software support short read assembling. Conversely, analyzing the long-read data is a challenging process.
Limited bioinformatics resources:
Existing bioinformatics tools, pipelines and software developed for short-read sequencing may not be directly applicable to studying long-read data. Thus, long-read sequencing data analysis is a computationally challenging, intensive and technically difficult process.
High error rate:
The present technique has a high error rate as it has to sequence a very long fragment. Hence, it creates problems in base calling and data analysis.
Low throughput:
Long-read sequencing is a low-throughput sequencing technique. It can generate fewer sequencing reads per run and therefore a limited number of samples can be sequenced.
Longer TAT:
The present technique has a longer TAT (Turn Around Time) as sequencing the longer fragments takes more time compared to short read sequencing.
High cost per base:
An important limitation of long-read sequencing is its higher cost per base. The complexity of the chemistry involved, coupled with expenses related to instruments, reagents, kits, and operational costs, contributes to an increased per-base sequencing cost.
Platform-specific bias:
Performing long-read sequencing can be challenging. The chemistry involved can differ significantly from one platform to another, leading to varying requirements and necessitating the use of distinct analysis tools for data interpretation and processing.
Requirement of high molecular weight DNA:
A critical limitation of long-read sequencing is its requirement of a high molecular weight DNA. This means that it may not be suitable for DNA samples with low molecular weight or limited quantities, posing a significant constraint on its applicability in diagnosis.
Related articles:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- What is NGS?- Definition, Principle, Steps, Chemistries, Advantages and Limitations
- De Novo Sequencing: Steps, Procedure, Advantages, Limitations and Applications
Wrapping up:
In conclusion, long-read sequencing isn’t a good choice for clinical studies and diagnosis, as for now! But it has the potential to penetrate the diagnostic market. The requirement of high molecular weight DNA and low throughput are the two major reasons, it can not be recognized well.
The extensive capabilities of long-read sequencing in investigating complex structural alterations and repetitive genomic regions offer significant promise for future research. The collaborative utilization of both long and short-read sequencing methodologies holds transformative potential for advancements in medical science and healthcare outcomes.
Sources:
Sequencing 101: Long-Read Sequencing By PacBio.
Long read sequencing by Genomic notes for clinicians.
Jain, M., Koren, S., Miga, K. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 36, 338–345 (2018). https://doi.org/10.1038/nbt.4060.
Payne A. et al. (2018) Whale watching with BulkVis: A graphical viewer for Oxford Nanopore bulk fast5 files. BioRxiv, 2018. https://doi.org/10.1101/312256.
Marx, V. Method of the year: long-read sequencing. Nat Methods 20, 6–11 (2023). https://doi.org/10.1038/s41592-022-01730-w.


