“When transposons are inserted into the host genome, it generates duplicate copies on both ends of the insert known as Target site duplication. Learn about the target site duplication and its significance in this article.”
Transposons, often known as transposable elements, these sequences are considered mobile genetic elements. TEs can move from one to another location in the genome thereby producing genomic variations.
Two types of transposons– retrotransposons and DNA transposons follow different mechanisms for transposition, however, the target site duplication is a crucial phenomenon using which transposons insert into the genome.
Let’s find out what target site duplication is and how it occurs.
We covered the entire series of transposons, you can read it here: Transposons.
Disclaimer: Information provided here is collected from peer-reviewed resources and re-presented in an understandable language. All the sources are enlisted at the end of the article.
Key Topics:
What is Target Site Duplication?
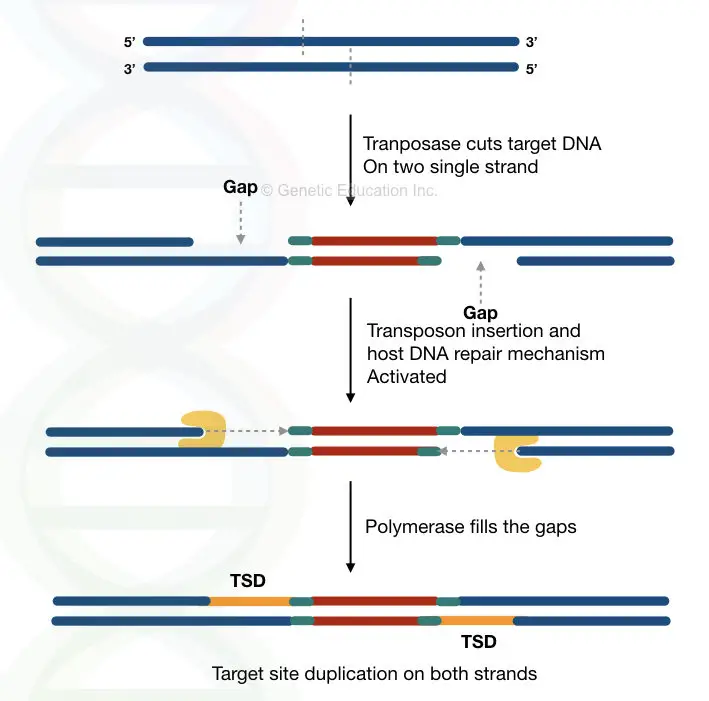
The target site, as the name suggests, is the target location in the host genome where the transposon will likely be inserted. Upon insertion, the transposon generates sticky ends which when filled, create sequence duplication.
The transposon insertion that produces copies of a sequence on both ends is known as target site duplication. Note that it occurs in the host genome, as a consequence of insertion. DNA transposons and some retrotransposons enter into the host genome by generating a target site duplication.
Henceforth, target site duplication is the hallmark feature of the transposons, we can say. Now coming to the point, how it occurs.
How does Target Site Duplication occur?
Enzyme transposase, encoded by the gene part of the transposon, catalyzes the entire reaction, starting from excision from the old site to insertion at the new location.
Therefore the entire random process and the rate of transposition depends on the transposase enzyme. In both, retrotransposition and DNA transposition, the excision process occurs using the same mechanism.
Now first, it finds the transposon terminal repeats, identifies it, cuts it from the location, creates a loop and removes it from that location.
Next, the enzyme recognizes the target location, which is a random process and settles there. Immediately, it introduces a cut into both single strands of the DNA and generates sticky ends (see the above image).
Sticky ends are different from blunt ends as they contain a few nucleotide overhangs. Afterward, it introduces the transposon into the target site. Note that transposition mostly occurs in the AT-rich regions but the theory lacks concrete data.
Now, the transposase protein inserts the transposon into the target site, but the overhang gaps remain unfilled. A cell’s natural DNA repair mechanism with the help of the DNA polymerase, fills the gap.
In the final step, DNA ligase ligate the sequences through phosphodiester bonds and completes the process. The hypothetical representation is given in the image and it shows how the duplication occurs.
Now, the transposon is inserted into the host genome and if it is present in the coding region, it mutates it and creates a new allele. Target site duplication occurs by the collective effort from both– the transposon machinery and the cell’s DNA repair mechanism.
Significance:
Now you may have a question, why does it occur? I have a one-line answer— to provide new alleles (variations). Here are some of the reasons why it occurs.
TSD leads to genomic rearrangements and new allelic combinations. Thus, it produces new traits or improves the existing ones and helps to produce variability.
Transposition can destabilize the genome, when a cell encounters such an event it immediately repairs it, just like the duplication event in this scenario and tries hard to stabilize the genome. Genomic instability can cause lethal consequences.
Recent studies suggest that TSD also influences gene expression. TSD duplication also regulates gene expression. The insertion of transposon and the creation of TDS can potentially influence a gene’s function and thereby regulate its expression. Largely, it down-regulates gene expression.
Target site duplication is a hallmark feature of transposable elements, as we said. Usually, such variations are lethal, but if it helps, they will remain, pass to consecutive generations and spread in the population. Therefore, transposon-mediated TDS provides variability in the population.
Wrapping up:
In conclusion, Target Site Duplication has positive and negative effects on the host genome. As the TDS process is random, the consequences that it produces are entirely random, unknown and unpredictable.
Nonetheless, the sole intention behind doing all these activities by nature is to create new allelic variations and produce diversity according to the requirement. Scientists are utilizing this hallmark genomic hotspot to understand the comprehensive mechanisms behind transposition.
I hope you like this article. Read other articles of this series as well.
Sources:
Linheiro RS, Bergman CM. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PLoS One. 2012;7(2):e30008. doi: 10.1371/journal.pone.0030008. Epub 2012 Feb 9. PMID: 22347367; PMCID: PMC3276498.


