“Whole genome and whole exome sequencing are two NGS platforms that can investigate the entire genome and coding sequences, respectively. Learn about the common and technical differences between WGS and WES.”
Our genome is made up of billions of nucleotides. While only 2% can translate into proteins, non-coding sequences regulate this entire process. So any mutation or alteration within coding and non-coding sequences has a significant impact on phenotypes.
Mutations eventually result in various diseases of complex genetic conditions. DNA sequencing can help to study and determine mutations or alterations within the coding regions or from the genome.
DNA Sequencing revolutionized the field of genetics by providing the power to understand sequence-level alterations. Put simply, sequencing helps to decode the genetic code and provides valuable information regarding our DNA.
The modern-day DNA sequencing platform known as Next-Generation Sequencing or NGS can sequence the entire genome consisting of 3.2 billion bps. Unfortunately, previous-generation sequencing lacked this much sequencing power.
However, it’s also difficult to sequence the entire genome in a single run even using NGS, scientists cut DNA into small fragments and sequence each fragment in a massively parallel fashion. In this article, we will discuss two of the most common and popular NGS platforms: whole-genome sequencing and whole-exome sequencing.
We will look at the differences between WGS and WES including the applications and process of working. This article will help research students gain knowledge about how these sequencing can help in different research modules as well as will help to make a choice for selecting the NGS platform.
We will first get clarity at genome and exome and then step-by-step get into the topic.
Stay tuned.
Common short forms:
| Short form | Full name |
| NGS | Next-Generation Sequencing |
| WGS | Whole-Genome Sequencing |
| WES | Whole-Exome Sequencing |
| SNP | Single Nucleotide Polymorphism |
| Indels | Insertions and deletions |
| SVs | Sequence variations |
| CVNs | Copy Number Variations |
| cDNA | Complementary DNA |
| PCR | Polymerase Chain Reaction |
| mRNA | Messenger RNA |
(We will use short-form WGS for Whole-Genome Sequencing and WES for Whole-Exome Sequencing throughout the article.)
Now, to understand it more precisely, let’s start with ‘Genome’ and ‘Exome’
Key Topics:
Genome vs Exome:
The entire genomic content of a cell is referred to as the genome while the coding portion of the genome is referred to as the exome.
Our genome is made up of coding and non-coding sequences consisting of exons, introns, transposons and pseudogenes, etc. The exome only contains exons or the coding sequences of the genome. So when we talk about the exome, only exons are covered.
Related articles: What are Pseudogenes?
Whole Genome vs Whole Exome:
The entire cellular DNA is known as the whole genome while the entire coding exon is collectively known as the exome. Hence, the technique used to sequence the Whole genome and whole exome is denoted as WGS and WES, respectively.
Now take a look at some of the differences.
Whole-Genome vs Whole-Exome Sequencing:
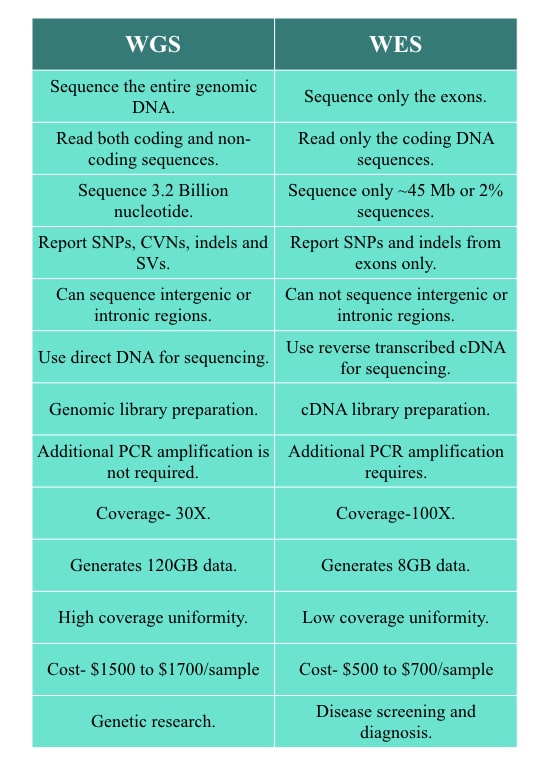
As the terminology depicts, WGS is a technique to sequence the entire genome of an organism while WES is a technique to sequence only the exome or coding part from the genome.
Alterations:
WGS: It can determine complete genome-wide alterations.
WES: It can determine alterations only from the coding regions. As coding regions participate in protein synthesis, such alterations are directly associated with various diseases.
WGS: It is a suitable technique for the detection of SNPs, indels, SVs and CNVs with high accuracy and sensitivity.
WES: It is a suitable technique for the detection of exonic SNPs and indels with high accuracy and sensitivity.
Coding vs non-coding:
WGS: It can determine the intronic or intergenic regions.
WES: It can not determine the intronic sequences.
WGS: It sequences coding, non-coding– Transposons, pseudogenes and mitochondrial DNA.
WES: It sequences only the exonic regions from the entire genome. Thus pre-sequence or reference information is required for exome sequencing.
Range of sequencing:
WGS: It sequences all the 3.2 billion nucleotides using a massively parallel sequencing technique with 99.9% sensitivity at 30X coverage.
WES: It sequences only 2% or (45 Mb) portion of the genome with 99.9% sensitivity and 100X coverage.
Process:
Technically both whole-genome and whole-exome sequencing uses the same next-generation sequencing platform but come up with different pre-processing setups. WGS: It requires direct DNA from the sample.
WES: It requires cDNA. cDNA is reverse transcribed from the mRNA. mRNA is synthesized only from the coding DNA or exons.
Thus, the steps in WGS are
- DNA extraction
- DNA fragmentation
- Library preparation
- Sequencing read
- Data analysis
While the steps in WES are
- mRNA isolation
- cDNA preparation
- PCR amplification
- cDNA Library Preparation
- Sequencing read
- Data analysis
This clearly shows that WGS requires a genomic library preparation scheme while WES requires a cDNA library preparation scheme.
Amplification:
WGS: It usually does not require any additional amplification step.
WES: It requires an additional reverse transcription PCR step to amplify the cDNA.
Coverage:
WGS: It can report alteration outside the exonic regions. This allows scientists to carefully investigate the role of non-coding alterations in disease development.
WES: It sequences only the exonic regions and thus can’t report alterations located outside of exons. However, it can report disease-associated variants more precisely.
Accuracy:
WGS: As it has to sequence a huge amount of data it has lower sequencing coverage and thus, has lower comparative accuracy.
WES: As it has to sequence a very small part of the genome it has higher sequencing coverage of up to 100X and thus, has very high accuracy comparatively.
Data processing:
WGS: It generates around 120GB of data with 30X coverage. Thus, it requires more computational power and bioinformatics resources.
WES: It generates around 8GB of data with 100X coverage. Thus, it requires less computation power and bioinformatics resources.
Coverage uniformity:
WGS: Despite lower coverage, WGS has high coverage uniformity.
WES: Despite having high coverage, WES has low coverage uniformity.
WGS: Due to lower sequencing coverage it can not capture rare variants more effectively than WES.
WES: Due to higher sequencing coverage (100X) it can capture rare variants more effectively. Thus, it directly finds and reports diseases associated genes and any rare variants from the exome.
Cost:
WGS: This technique is costly as it requires higher sequencing power, storage, processing and analysis capacity.
WES: This technique is comparatively cheaper as it requires less sequencing power, storage, processing and analysis capacity.
Cost per sample:
WGS: As per the recent data, the cost of whole-genome sequencing including library preparation is approximately $15,000 to $1700 per sample.
WES: The cost of whole-exome sequencing including cDNA Library preparation is approximately $500 to $700. Which is still very low compared to WGS.
Reads:
WGS: It can read both long and short reads effectively.
WES: It can only read short reads effectively.
Application:
WGS: It can identify novel and rare variants associated with a disease Which helps to investigate complex genetic or disease conditions.
WES: It can study genes associated with Mendelian and other genetic disorders and thus, can be utilized in clinical setups.
Further reading: What is Sequencing reads?
Wrapping up:
Whole-genome sequencing is an excellent tool for genetic research and finding novel disease-associated variants— SNPs, indels and CNVs while whole-exome sequencing is a perfect tool for clinical analysis and diagnosis as it readily helps to study disease-associated known genes and related alterations.
Due to the low cost and fast processing, the clear winner here is whole exome sequencing for healthcare. However, WGS can give far more data than WES. I hope you like this article. If so please share it and bookmark the page.

