DNA extraction is a type of nucleic acid separation technique to isolate DNA from a biological sample. Many different DNA extraction methods scientists use. Let’s discuss 10 standard DNA extraction methods.
Download this article as an ebook: DNA Extraction Methods pdf.
Molecular genetic techniques highly rely on pure DNA or RNA. For example, techniques like PCR, sequencing, microarray, hybridization and restriction digestion require DNA. Studies demonstrate that pure DNA certainly warranties the success of the assay.
Henceforth, DNA extraction is important.
Genomic DNA is located in the cell nucleus. So to get DNA, we need to break a cell membrane or cell wall, and a nuclear envelope. However, it is indeed a difficult task. Because it requires a set of different chemicals and solutions, and physical techniques.
In addition, diverse cell structures often create a problem in this scenario. For example, the plant cell has a rigid cell wall that’s why DNA extraction from plant cells is difficult too.
“One size doesn’t suit all.”
Every tissue type needs a differential DNA extraction setup, thus, it makes overall things difficult for research students and scientists. Factors that cause problems in DNA extraction are,
- Differential cell composition.
- Different tissue types.
- The difference in genomic content.
- Chemicals used in the process.
- Solution preparation.
- Type of physical technique used.
- And many more…
So it is important to broadly understand the concept, requirements and process of different types of DNA extraction methods. I had my own research lab and I and my team had optimized many DNA extraction techniques.
So, I know a bit more about DNA extraction. I have published a couple of research works as well (that you can read on my profile page). In this article, I will explain various DNA extraction methods, concepts, and chemicals used and give a broad overview.
Stay tuned.
Key Topics:
What is DNA extraction?
The simplest explanation of DNA extraction is, “extracting DNA from a cell”. It is a type of nucleic acid extraction technique that only isolates the DNA. First, check out the short history.
History of DNA extraction
The first DNA extraction attempt was performed by Friedrich Miescher in 1869. He isolated the cell material and named it the “nuclei” later on his student named it, a “nucleic acid”. Although Miescher F accidentally developed a method for nucleic acid isolation, he was not sure whether what he isolated was DNA or not.
Later on, in 1958 Meselson and Stahl developed a full-function protocol for DNA extraction. The density gradient centrifugation protocol was the first protocol described for isolating DNA from E.coli bacteria.
In 1988 Miller et al. proposed the use of proteinase K in DNA extraction, nonetheless, Lahiri and Nurnberger (1991) used it effectively along with the Nonidet P40 and SDS in extraction.
Sambrook J and Russell D proposed one of the most popular and anticipated DNA extraction techniques, Phenol-chloroform and isoamyl alcohol. The PCI method is a more powerful, accurate and high-yield method and is still used in recent times.
Definition of DNA extraction:
“Isolating DNA by disrupting cell wall/cell membrane and a nuclear membrane is called a DNA extraction”.
“Isolation of DNA by breaking the cell membrane and nuclear membrane with the help of chemicals, enzymes or physical disruptions is defined as a DNA extraction”.
Or simply,
“Isolation of DNA from the rest of the cell components is known as DNA extraction.”
Principle of DNA extraction:
To effectively understand the concept, we have to divide the principle into three different steps. Cell wall/membrane and nuclear membrane lysis, stabilizing DNA, precipitation and washing.
To isolate DNA, we first have to break the cell membrane and nuclear membrane which comprises, roughly, glycoproteins and lipids. Heat, detergents and enzymes can do this job effectively.
Once we get the DNA, next we have to remove the contaminant without compromising its stability. Chemicals like EDTA and NaCl prevent DNA damage from DNase and protect it. Whereas centrifugation can effectively remove major larger cell debris from the DNA.
The pure nucleic acid is precipitated into a visible form using alcohol. Isopropanol is the first choice for precipitation. Washing with alcohol removes other protein debris from the DNA and gives us pure DNA.
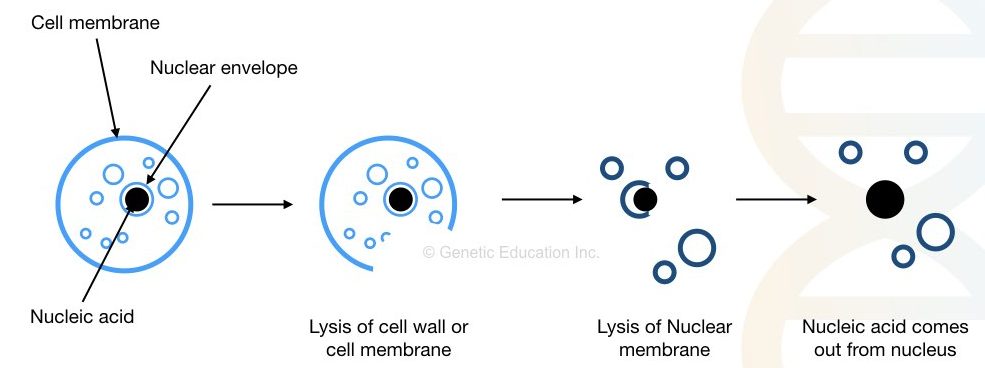
Put simply the principle is as followed,
“Removal of the cell membrane and nuclear envelope, stabilizing DNA from DNase, removal of cell debris, precipitation and washing.”
If you remembered, we have stated a problem of “difference in the composition of the cell.” This must be discussed first before going ahead because it clarifies many things regarding DNA extraction.
Animals have a smooth cell membrane, plants have a hard, rigid and rough cell wall while bacteria have a smooth cell wall. Therefore, each needs a different DNA extraction setup.
Cell with a smooth cell wall:
Bacteria commonly have a smooth cell wall. It is so rigid that gentle heating at 60 to 70°C can break it. Such a sample can directly be used in PCR or by adding an extra heating step things can be managed. For example, TB-PCR, in which we add the sample directly into the PCR and it works. However, the use of a lysis buffer increases the purity and yield of DNA, greatly.
Cell with a hard cell wall:
A hard, rigid and polysaccharide-rich cell wall is a unique property of a plant cell. Common physical techniques like heating and grinding and chemical techniques like the use of lysis buffers even fail to isolate DNA.
Moreover, the presence of secondary metabolites makes overall things more difficult and oftentimes, students are stuck here. We have written a dedicated article on this, the link is given above.
The use of liquid nitrogen, tissue homogenization, and chemicals like CTAB, PVP and SDS can effectively isolate DNA from plant cells.
Cell with cell membrane:
Animal cells lack a cell wall and only possess a smooth membrane. Using standard isolation chemicals or buffers and techniques like heating, one can effectively isolate nucleic acid. PCI DNA extraction and proteinase K-based methods are two commonly employed techniques for isolating DNA from animal cells.
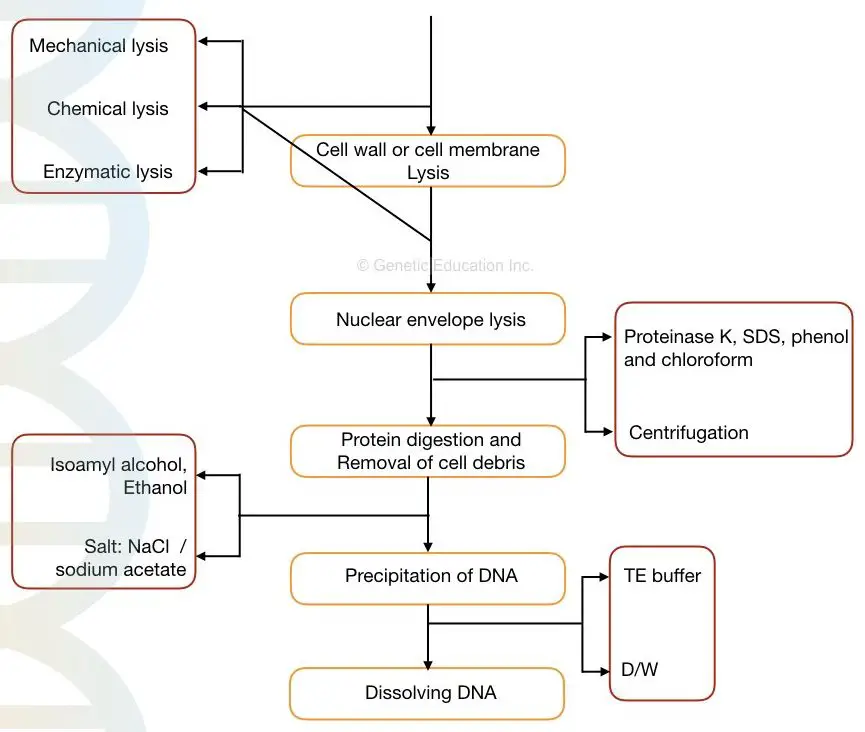
Steps in DNA extraction:
- Cell membrane or cell wall lysis
- Nuclear envelope lysis
- Removal of contaminant
- Stabilizing DNA
- DNA Precipitation
- DNA washing and elution
Chemicals used in DNA extraction:
Common DNA extraction chemicals and their roles are explained here in the table.
| Chemical | Function |
| Tris | Control the pH, interacts with lipopolysaccharides and increases permeability, and lysis cell membrane. |
| EDTA | Works as a chelating agent, blocking the cofactor requirement of the DNase enzyme, thereby preventing DNA from degradation. |
| SDS (Sodium dodecyl sulfate) | Anionic detergent that digests nuclear and cell membrane proteins. |
| NaCl | Neutralizes the negative charge of DNA and stabilizes and protects it. |
| MgCl2 | MgCl2 is another agent that protects and stabilizes the DNA by blocking the negative charge of lipoproteins. |
| Phenol | Precipitates protein impurities. |
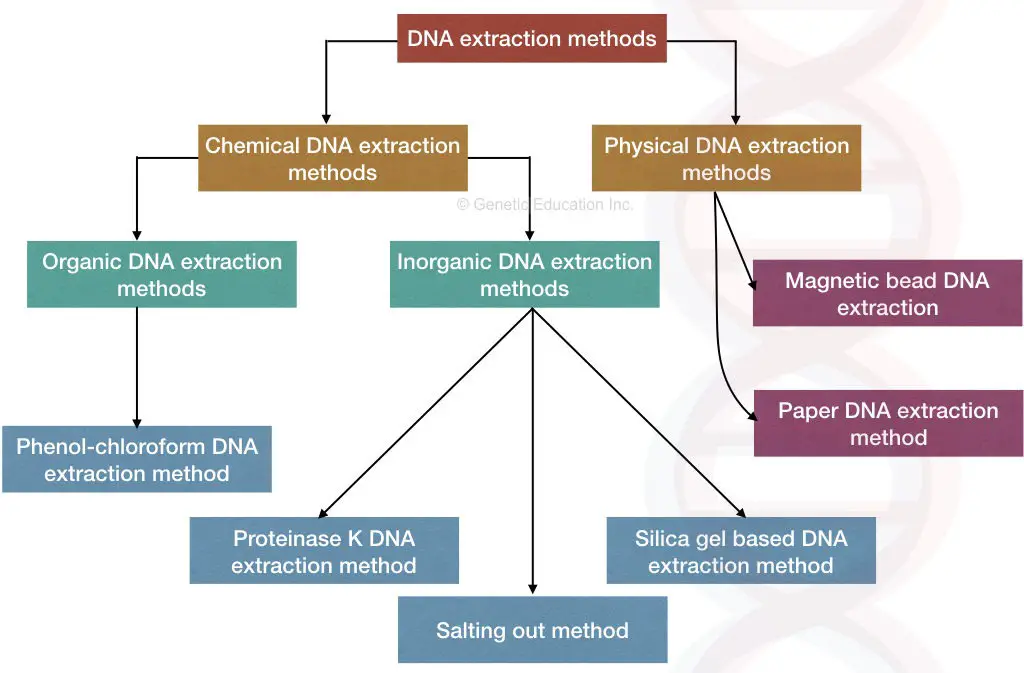
DNA extraction methods:
The DNA extraction methods are broadly categorized into chemical and mechanical methods, however, the classification is based on the primary technique implies, and the rest of the steps and chemicals sometimes remain the same.
Chemical DNA extraction methods:
- Organic DNA extraction
- Phenol-chloroform and isoamyl alcohol
- Inorganic DNA extraction
- Proteinase K DNA extraction
- CTAB DNA extraction
- SDS DNA extraction
- Salting out method
- Silica-gel-based techniques
Physical DNA extraction methods
- Magnetic bead DNA extraction
- Paper DNA extraction
Chemical DNA extraction:
Chemical DNA extraction is also known as liquid or solution-based DNA extraction. This group of techniques highly relies on different chemicals. Thus extensive chemical preparation is required.
The organic solvent-based separation uses organic chemicals, for example, phenol and chloroform. It involves risk and may harm us. The inorganic solvent-based techniques include other chemicals like SDS, CTAB, NaCl and MgCl2. These groups of techniques are safer to use.
Liquid-liquid DNA extraction:
One of the commonest methods for nucleic acid extraction is liquid-liquid extraction. Here only solutions prepared by various chemical compositions are used for extraction. hence, it mainly relies on lysis buffer preparation.
The overall success of the experiment depends on how effectively the lysis buffer is prepared.
Lysis buffer is often prepared in one or two solutions as it uses many chemicals. Common chemicals for liquid-liquid DNA extraction are Phenol, chloroform, isoamyl alcohol, SDS, CTAB, Triton X100, Tris, EDTA, MgCl2, NaCl, PVP, beta-mercaptoethanol and other detergents.
These methods commonly require centrifugation for separation. Common liquid-liquid DNA extraction methods are,
- Phenol, chloroform and isoamyl alcohol DNA extraction
- CTAB DNA extraction
- SDS DNA extraction
Solid-liquid phase DNA extraction:
Spin-column and magnetic bead-based separation techniques are solid-liquid phase DNA extraction that uses both liquid lysis buffer and solid phases like silica gel or magnetic beads.
These groups of techniques are safe, fast and easy to perform but at the same time are costly.
How to do mechanical lysis?
Take plant tissue and grind it with a mortar and pestle. Grind it well, and add liquid nitrogen carefully and again grind it even harder until the tissue becomes powder. Finally, add lysis buffer or DNA extraction buffer. Grind it till it becomes homogeneous. The tissue is ready for DNA extraction.
Additionally, we can add an enzyme to this homogenized solution and incubate it for an hour to get good results. The combination of mechanical-chemical and enzymatic lysis helps to extract good quality DNA from the plant cell. This combination most probably gives the best result.

10 Different DNA extraction methods:
In this section, we will discuss common individual extraction methods and their recognition.
1. DNA extraction by chromatography:
- Type: Physical separation technique
- Subtype: Solvent-based DNA extraction
- Purity: Decent (nearly ~1.79 to 1.88)
- Yield: Decent
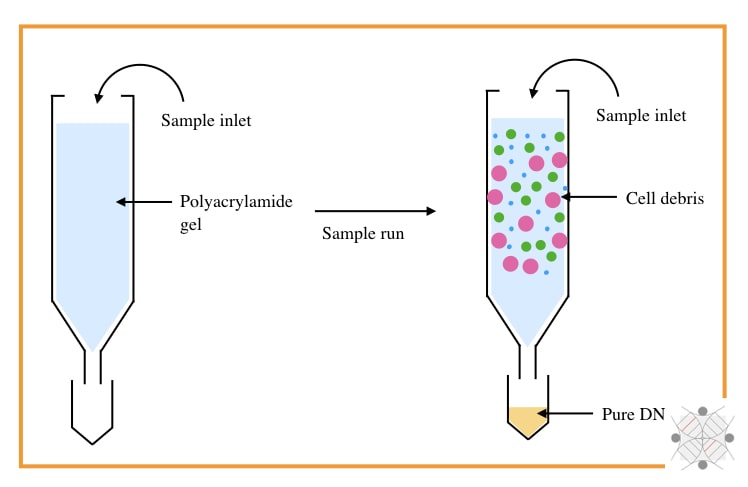
Chromatography-based nucleic acid separation, however, is not used in recent times but was part of the history of DNA extraction. SEC (Size exclusion chromatography) and IEC (Ion exchange chromatography) were certainly popular for separation.
SEC is a size-dependent technique that uses the size of the molecule to perform the separation. A column filled with agarose, polyacrylamide or dextran, only allows the migration of DNA from smaller gel pours.
The rest of the debris like protein and lipids remains in the column while DNA elutes out. However, this technique can’t remove RNA, effectively.
The IEC allows effective isolation of DNA without RNA traces. Ions present in the column selectively bind with the DNA, remove other contaminants like proteins, lipids and RNA, and separate DNA in the next round by pH change.
Ion exchange is more powerful than conventional chromatography. Both techniques give lower yields.
| Advantages | Disadvantages |
| Easy to perform and comparatively rapid. | Low yield and quality. |
2. DNA extraction by CsCl density gradient centrifugation:
- Type: Physical separation technique
- Subtype: density gradient centrifugation
- Purity: Not good
- Yield: Not good
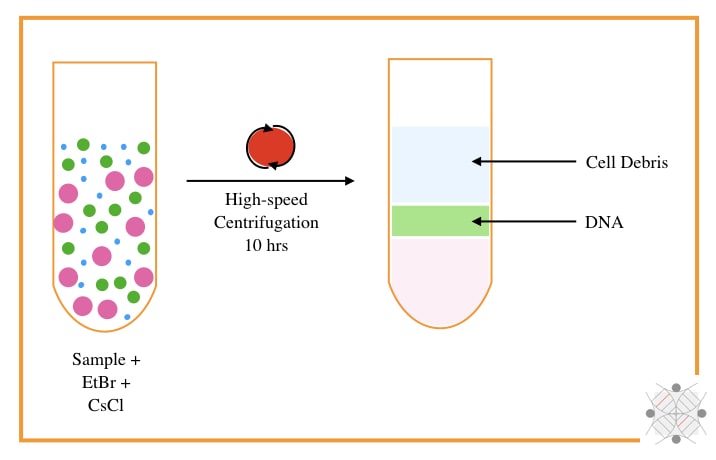
The CsCl-Density gradient centrifugation using EtBr was developed by Meselson M, Stahl F and Vinograd J in 1957. The technique originally requires a high-speed centrifugation system, CsCl and EtBr.
In principle, when the DNA reaches the density of the CsCl, at an isopycnic point, it will appear. The sample is mixed with the CsCl and centrifuged at 10,000 to 15,000 rpm for 8 to 10 hrs.
The EtBr used during the process intercalates with the DNA and gives a band of DNA under UV light. As the intercalating capacity of EtBr is very high for non-supercoiled DNA, it can effectively isolate bacteria and plasmid circular DNA.
The particular application of the CsCl centrifugation method is that it can isolate circular and non-supercoiled bacterial and plasmid DNA, however, it has several limitations too.
The present technique is complex, tedious, time-consuming and costly. The use of EtBr and UV light makes it more unsafe. In addition, the yield and purity are also very low.
| Advantages | Disadvantages |
| Can isolate non-supercoiled, circular, plasmid and bacterial DNA. | Tedious, time-consuming, unsafe, low yield and quality. |
3. Phenol-chloroform and isoamyl alcohol:
- Type: Chemical/liquid separation technique
- Subtype: Solution-based (liquid-liquid) organic DNA extraction
- Purity: Excellent (~1.80)
- Yield: Excellent
- Sample: Animal, plant, fungi, bacteria or even plasmid
- Tissue type: Blood, saliva, solid tissue, plant leaf or root, cell culture, pleural fluid, skin or even hair.
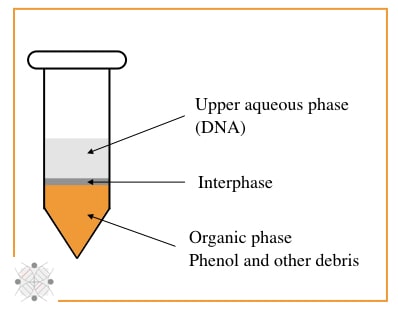
My personal favorite and trusted DNA extraction technique, is PCI, although it uses unsafe chemicals. The PCI method relies on the principle of liquid-liquid DNA extraction and is categorized into organic separation methods.
Organic solvents phenol and chloroform, and isoamyl alcohol are major players here. In principle, the phenol digest proteins, isoamyl alcohol separate nucleic acid and chloroform reduce the foaming between interphase.
In the last step, by isopropanol precipitation, the DNA can be isolated from biological material. The main advantage of the PCI is that it can isolate DNA from nearly all tissue types. Furthermore, it can even isolate RNA when used with guanidine thiocyanate.
It works for animal, plant and bacterial cells and tissues like blood, solid tissue, saliva and other body fluids. The quality and yield of DNA obtained by PCI are also excellent.
However, on the downside, it is unsafe.
Phenol is volatile and can burn the skill, the chloroform can faint us. Thus care and experience are required to perform it. Notedly, it requires other chemicals like Tris, EDTA, MgCl2, KCl, NaCl, and sometimes SDS and isoamyl alcohol.
Read more: Phenol, Chloroform and Isoamyl alcohol DNA extraction.
| Advantages | Disadvantages |
| The quality and yield of the extract are excellent.The technique is cheap, easy and reliable | Unsafe and health hazardous. Required extensive and tedious chemical preparation. It is time-consuming and requires training and/or experience to prepare and handle chemicals. |
4. CTAB DNA extraction:
- Type: Chemical/liquid separation technique
- Subtype: Solution-based organic DNA extraction
- Category: Liquid-liquid separation
- Purity: Excellent (~1.80)
- Yield: Excellent
- Sample: Plant sample
- Tissue type: Plant leaf, bark, dead plant parts and root.
In 1990, Doyle introduced the CTAB method for plant DNA isolation. Technically, it needs a low ionic strength buffer and pH >7 (alkali pH).
CTAB- Cetyl Trimethyl Ammonium Bromide is a chemical often used in DNA extraction. The CTAB-based DNA extraction is a specially prepared liquid-liquid and solution-based extraction method for plant DNA extraction.
It is also important to know that to improve the efficiency of the protocol other chemicals like SDS, Beta-mercaptoethanol, PVP and Triton X 100 are also combined. CTAB and SDS remove solid glycoproteins and the rest of the chemicals remove polyphenolic compounds.
Evidently, enzymes like cellulase and pectinase, etc are often combined with the technique to remove hard polysaccharides of the plant cell wall.
2 to 5% CTAB and other ingredients are prepared in the form of lysis buffer and purified using the PCI or alcohol methods. On the positive side, it can effectively isolate DNA from hard materials like plants.
Read more: CTAB DNA extraction buffer for plant DNA extraction.
| Advantages | Disadvantages |
| The CTAB buffer removes polysaccharides and polyphenols effectively. Gives excellent yield for plant DNA. | It required extensive chemical preparation. It is a time consuming and tedious method. It also requires additional techniques like tissue homogenization and the use of liquid nitrogen. |
5. Proteinase K DNA extraction:
- Type: Chemical DNA extraction
- Subtype: Enzymatic technique
- Purity: Excellent (~1.80)
- Yield: Excellent
- Sample: any biological
In 1974, Ebeling et al. discovered the serine protease- proteinase K. It is one of the most common protease enzymes used in DNA extraction. Technically, it governs an enzymatic catalytic reaction to neutralize, unfold and digest nuclear and cellular proteins.
Studies show that when combined with the SDS method, it removes all forms of protein from the sample. However, a separate cell lysis solution is often required. It works for animals, plants, bacteria and even fungal samples.
Each sample requires 10 to 20 mg/ mL of proteinase and incubation for 1 to 24 hrs at 60 to 65°C temperature.
To know more, you can read our protocol on proteinase K: Proteinase K DNA extraction method.
| Advantages | Disadvantages |
| Easy to use, provides high purity and yield. Safe to use, accurate and rapid. Can be used for any sample. Can be combined with any protocol. Require a small sample volume. | A bit costly, additional SOP to store and process proteinase K is needed. |
6. Spin-column DNA extraction:
- Type: Chemical DNA extraction
- Subtype: Solid-liquid phase DNA extraction method
- Purity: Excellent (~1.80)
- Yield: Good
- Sample: Any biological sample
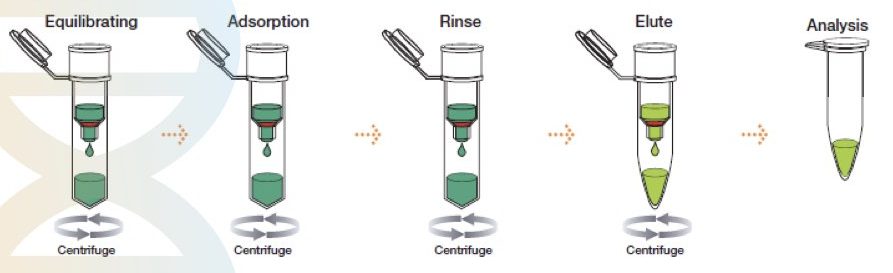
The method was first described by McCormick in 1989. However, the idea was developed in 1979, when silica was used in DNA purification by Vogelstein.
The spin-column DNA extraction technique has evolved recently and is the most advanced, rapid, effective, easy to use and accurate separation technique. It is, however, a chemical method of DNA extraction but works on the principle of solid-phase separation.
A silica gel as a solid phase is immobilized in a tube. A specially prepared lysis buffer is used to lyse the sample and subsequently allow it to separate on a solid phase. Centrifugation first removes all the debris by protecting the DNA and elutes DNA in the last step by changing the pH of the solution. Such spin-columns and lysis buffer are nowadays commercially available.
This technique can isolate DNA from any biological sample and tissue. However, it is difficult to isolate DNA from plants even using the spin-column technique. Notedly, different columns are now available depending upon the sample type.
| Advantages | Disadvantages |
| Fast, accurate, easy to perform and safe. Isolate high-quality and yield DNA. Doesn’t require precipitation. Can process any sample. Works for small volume samples. | Costly. Lacks optimizations. The yield remains comparatively low but good. |
7. Magnetic bead-based DNA extraction:
- Type: Physical DNA extraction
- Subtype: Solid phase DNA extraction method
- Purity: Excellent (~1.80)
- Yield: Excellent
- Sample: Any biological sample
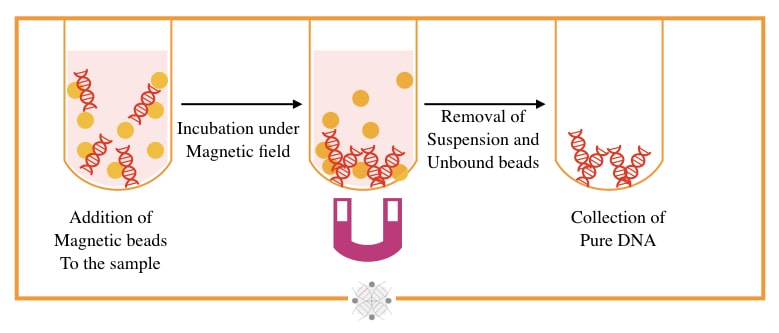
The magnetic bead-based technique is a state-of-art, speedy and most accurate technique for the separation of DNA, originally developed by Hawkins T in 1998. Put simply, under the magnetic field, the bead-bound DNA extracts.
Magnetically charged beads made up of sulfate and hydroxyl groups, and DNA binding antibodies or polymers, are used to separate DNA. Beads are mixed with DNA and allowed to separate in a high magnetic field.
The bead-bound DNA will remain on the bottom while all other debris comes out in the supernatant. The beads are washed and DNA eluted. In the final step, the DNA detached from the beads by heating at 60 to 65°C temperature.
The present technique is so speedy and completes within 15 to 20 minutes, and thus can handle a higher sample load. The yield and quality of the final elute are also very good. However, it needs an additional instrumental setup.
| Advantages | Disadvantages |
| Fast, accurate, safe, reliable and can handle huge sample loads. Doesn’t require centrifugation and extensive chemical preparation. Automated. | Constantly. Additional magnetic instrument setup is required. |
8. Paper DNA extraction:
- Type: Physical DNA extraction
- Subtype: – Solid-liquid DNA extraction
- Purity: -N/A
- Yield: -N/A
- Sample: – N/A
I once used paper for DNA extraction from a blood sample. The presentative claimed that it is a fast and reliable isolation technique. But probably I don’t remember if it was a good one or not. But we never used it after, so surely it was a bad one.
Anyway, Let us see how it works.
When a drop of sample is applied to the cellulose paper, treated with a washing buffer and resuspended into the elution buffer, DNA can be extracted, however, perhaps all the magic would be in the composition of lysis and elution buffer.
The manufacturer claims that paper-based DNA extraction is a fast, accurate and high-yield extraction technique and would be the future of nucleic acid extraction. Nonetheless, I didn’t find it interesting when I was working in a lab.
| Advantages | Disadvantages |
| Fast, easy to use, cost-effective and safe. | Not as effective as other assays. Can’t be used for getting DNA from the least sample. |

9. Anionic resins DNA extraction:
- Type: chemical DNA extraction.
- Subtype: – Liquid-liquid DNA extraction
- Purity: – not good
- Yield: -decent
- Sample: – Any
Unlike RNA extraction in which RNase inactivation is an extensive goal, we usually, less care about the DNase during DNA extraction. The reason is clear, DNase isn’t as common as RNase.
Chelex protects DNA from DNase, thereby increasing the yield of isolated. In 1991, Walsh et al. first time used Chelex anionic resins in DNA extraction. However, Hui et al. patented the process in 2011.
Simultaneous use of anionic resins and proteinase K will chelate the metal ion for DNase and digest the proteins of a cell. Centrifugation recovers the DNA. Common anionic resins are styrene divinylbenzene and diethylaminoethyl.
Put simply, the process completes in three steps: add chelex 100 and proteinase K to the sample, boil it, centrifuge it and collect the nucleic acid.
| Advantages | Disadvantages |
| Rapid, cost-effective, safe and easy to use. | Can’t remove all the contaminants, is inaccurate and isn’t suitable for many downstream processes. |
10. Guanidine thiocyanate:
Type: chemical DNA extraction.
Subtype: – Liquid-liquid DNA extraction
Purity: – not good
Yield: -Excellent
Sample: – NA
Guanidine thiocyanate is a chaotropic salt and a denaturing agent often used in nucleic acid extraction. It can effectively degrade various proteins and is employed along with phenol, chloroform and isoamyl alcohol.
However, as many excellent protocols are available for DNA extraction, and the tendency of Guanidine thiocyanate to deactivate RNase, it is routinely used in RNA extraction. Notedly, it is one of the ingredients of lysis buffer formulation.
| Advantages | Disadvantages |
| Good chaotropic and denaturing. | Routinely used for RNA extraction. |
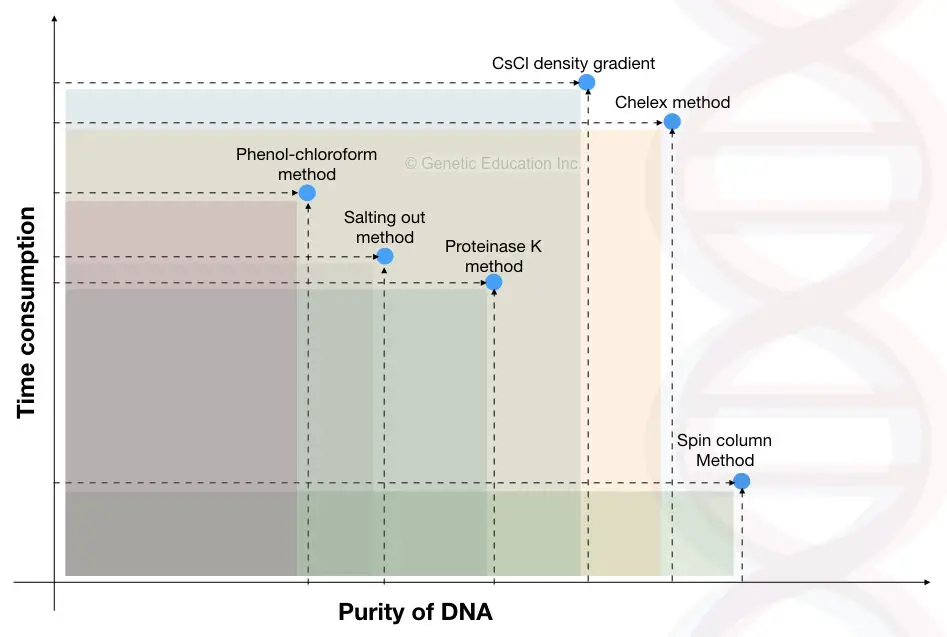
Suggestions from my personal experience:
I isolated DNA 10,000 or more times in my life, worked with many of these listed methods and optimized so many protocols. From my personal experience, let me tell you by far PCI, CTAB and spin-column-based DNA extraction is so good.
However, I still do not recommend the spin-column as it is a costly and low-yield technique. Using a manual method enables us with the power to optimize the chemical composition and lysis buffer recipe.
For example, if you have a ready-made lysis buffer and you know it doesn’t include SDS, if you get protein contamination, you can add SDS, additionally as per the requirements. I have prepared many combinations of lysis buffers for different sample types.
Get access to it by reading this article: How to Prepare Lysis Buffer for Different Types of DNA Extraction Methods?
To get success, weigh chemicals accurately and prepare solutions as per the protocol. Do all the steps as per the instructions.
Chemicals like phenol, chloroform and EtBr are unsafe and can harm our health so wear gloves, eye protectors, lab-coat and follow safety precautions.
If you want high yield DNA, use manual methods, if you want high pure DNA, faster use ready-to-use kits. If you want to sharpen your DNA extraction skill, you can buy our ebook: from DNA extraction to PCR.
Read more:
Wrapping up:
Trust me no, ready-to-use kits are powerful enough to extract good DNA, especially from the samples like plants. Every time, for every sample, you have to purchase a new kit. So it is a better idea to develop our own recipe.
Sometimes, your recipe for a lysis buffer even does better than kits. Once you purchase the utilities, you can work for 2 to 3 years. So it is genuinely cost-effective.
Moreover, working on manual protocols will help you to understand the basic principle behind how things work. That would become very important in the long run. So I personally advise you to start with manual things. Do not directly shift on kits.
FAQs on DNA extraction:
Download this article as an ebook: DNA Extraction Methods pdf.
Sources:
- Eslami G, Khalatbari-Limaki S, Ehrampoush MH, Gholamrezaei M, Hajimohammadi B, Oryan A. Comparison of Three Different DNA Extraction Methods for Linguatula Serrata as a Food-Borne Pathogen. Iran J Parasitol. 2017;12(2):236–242.
- Dilhari et al. “Evaluation of the impact of six different DNA extraction methods for the representation of the microbial community associated with human chronic wound infections using a gel-based DNA profiling method. ”AMB Expr (2017) 7:179. DOI 10.1186/s13568-017-0477-z.
- Kelly M. Elkins. “Chapter 4: DNA extraction.” Forensic DNA biology (2013): 39-52.



the best article about DNA extraction in very simple words.. Thank you
Thank you for Genetic education team
Pingback: DNA EXTRACTION LAB – Site Title
The best article I ever red about DNA extraction
Many thanks
Thank you Dr Shimaa
This has been the most helpful article I’ve read on DNA extracion. It really helped me understand the bigger picture.
Could I have the references?
My email is [email protected]
amazing article.! thanks you a lot but i who are the authors of the article?I must write references,so i should know author and also year.
plzzz would u send me refrences?
[email protected] on this address
References are there at the end of the article
I’m a research student who is interested in this area. Is this a good topic for an computer Science student to start a research? Can I get some advice from you?
Is this a good topic for my master program? I’m a Computer science student. If this is good, Can you give me some information on how to start my research?
Thank you in advanced.
Mail us your query
Could you please provide an email address? or Can I send an email to this address “[email protected]”?
yes, it’s our official email id.
Best ever data am going to dtp it then print…sir can u please give any dtp type material so that I can print it?
Really informative and helpful. A great “thank you” to all who made it possible.
This is an amazing article but I need the references of it
Hi, first of all, thanks for creating this blog, really helpful.
This article was so informative about the methods of DNA extraction.. Helped a lot..:)
Thank you Rashmi Vijay
This article is really nice but there are no references. How do i get the references?
Give me your email address i will send you the references.
if you don’t mind, can I have one?
[email protected]
Where are the references of this article ?
You can search it on the web. The article is written from our own experiences and some extra research.