“A hairpin-shaped, single-stranded, sequence-specific real-time PCR probe is known as Molecular Beacon having a higher sensitivity and specificity.”
The present article is more technical, including in-depth technical things related to the RT- PCR. If you are a PCR geek, stay on this article or read some of the basic articles on the present topic.
“Probe” is an important molecular for Real-time PCR. A type of PCR, Real-time, or RT-PCR not only amplifies the nucleic acid but also measures the amount. Some of the common probes employed are TaqMan probes, scorpion probes and molecular beacon probes.
Among all, the TaqMan probes and probe chemistry are widely used and most trusted. However, sometimes, the reaction needs some other type of probes. Let me tell you that the probes are much like the PCR primers, single-stranded, template-specific nucleic acid sequences.
Probes are fluorescently labeled whereas primers aren’t.
The present article will more discuss the use and role of molecular beacon probes in real-time PCR and other applications. But before that, if you want to learn more on the present topic, you can read this article.
Key points:
-
- A brief introduction to real-time PCR
- What is the molecular beacon?
- Structure of molecular beacons
- Designing of molecular beacons
- Importance of molecular beacons
- Application in genomics
- Conclusion
Key Topics:
A brief introduction to real-time PCR:
The real-time PCR or the quantitative real-time PCR is a method in which we can quantify the nucleic acid (DNA or RNA), efficiently. It can also measure gene expression.
The technique relies on Fluorescence chemistry, here the fluorescent-labeled oligo binds at the complementary location on a template. Correct hybridization emits fluorescence which is detected by the PCR detector. The amount of fluorescence emitted is directly proportional to the amount of template in the sample.
Dye-based method and Probe-based hybridization methods are two commonly available techniques. In the dye-based method, the colored dye binds to the template nucleic acid and gives results while in the probe-based method, a complementary probe binds to the nucleic acid and gives results.
Usually, the probe-based technique is more specific and efficient and therefore used widely in diagnostics. As we said, three common probes are
-
-
- Linear probes
- Molecular beacons
- Scorpion probes
-
Here in the present article, we will only discuss the molecular beacon probes.
What is a “molecular beacon” Probe?
Sometimes a reaction needs a specialized probe set up to increase the specificity. The molecular beacons are the specialized TaqMan probes used in Real-time PCR to increase the specificity of the reaction.
It is a single-stranded oligonucleotide hairpin structure made up of 25 to 30 nucleotides. In which the 15 to 20 nucleotide present into the middle-loop is complementary to the sequence of our interest. Whereas the sequences present on both arms are complementary to each other.
So structurally, as the sequences on both arms are complementary, they bind and create a loop. See the structure explained below.
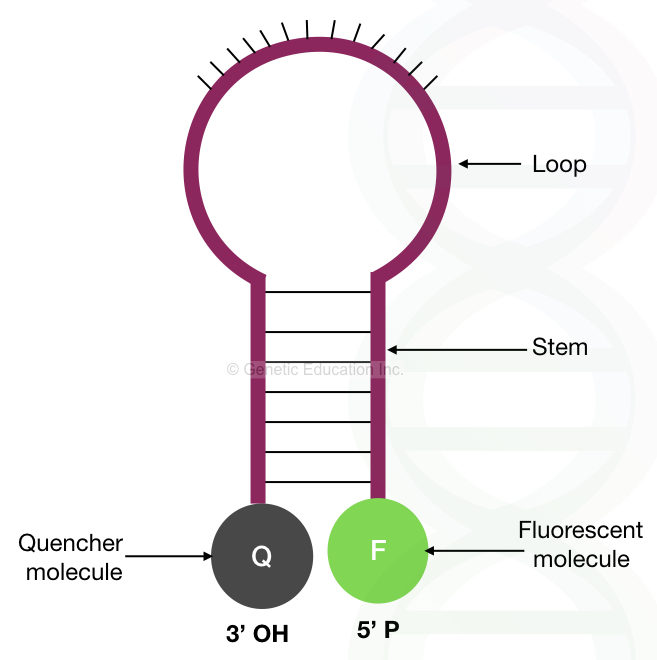
Structure:
The molecular beacons consist of 4 parts. A stem, loop, 3′ end and the 5′ end.
Stem:
The stem consists of 7 to 9 nucleotides that are complementary to each other. The nucleotides in the stem remain bounded with each other if the beacon is in a relaxed state.
Loop:
The loop is the central part of the molecular beacons, which is very important as it complements the target sequence. The central loop part contains 15 to 20 nucleotides, complementary to the sequence of our interest.
3′ end:
The 3′ end of the molecular beacons contains the quencher dye. This quencher dye on the 3′ end is in close proximity with the 5′ end, prevent the emission of fluorescence when relaxed.
5′ end:
The 5′ -OH end of the probe contains the fluorescent dye. Once the beacon binds to the complementary sequence, the fluorescent dye is unquenched, released and emits the fluorescence.
Most of the reactions commonly need a linear TaqMan probe then you may wonder why we need the molecular beacon probes.
Importance of molecular beacon:
The real-time PCR is mainly used to quantify templates and hence used in sensitive experiments or diagnostics. Any mistake during the assay isn’t tolerable. But the non-specific binding creates probes in PCR and real-time PCR. Meaning, the probe binds to sequences other than its complementary one
Now if TaqMan probes are so successful, then what is the problem with them?
The linear TaqMan probes are 18 to 20 nucleotide long sequences on which one end contains the fluorescent dye while another end contains the quencher dye. If it finds some of the complementary bases, it hybridized, the fluorochrome unquenched and releases the fluorescence.
So using a linear probe in the real-time endpoint or real-time PCR assay is not a good choice always.
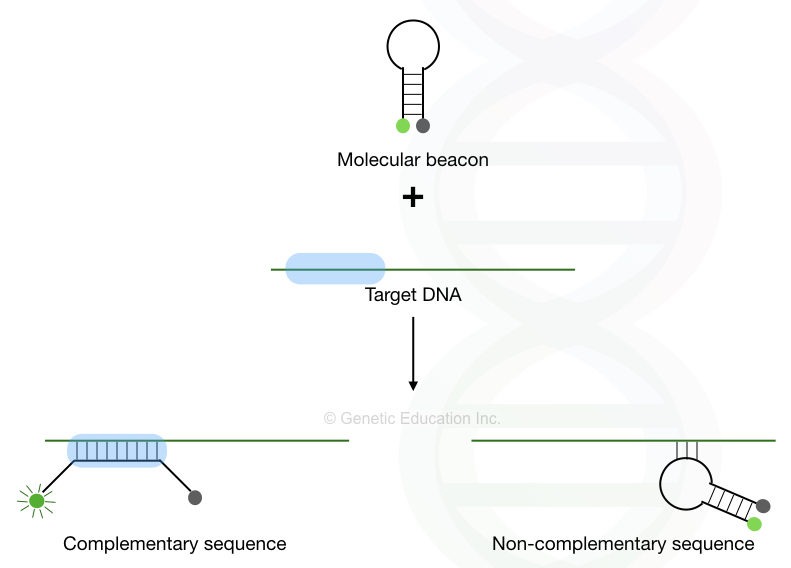
The molecular beacons are specially designed probes on which if it is not bound to any sequences, the majority of its energy is saved (remains in the hairpin state- relaxed state). But once it bound to its complementary sequence or in the presence of the complementary sequence the beacons unfold.
The fluorochrome no longer quenched. It is released and emits fluorescence. The detailed stepwise explanation of the mechanism of molecular beacons’ action is shown in the figure below.
![]()
The same reaction occurs on every template, only when the probe finds its exact complementary sequence, if binds, unquenches and releases fluorescence. DNA templates are measured in this fashion. After each PCR cycle, more probes are hybridized more fluorescence is emitted and more templates are quantified.
The entire mechanism of the molecular beacons is based on the stability of the hybrid formed during the amplification (DNA: DNA hybrid or RNR: RNR hybrid).
The stability of the perfectly matched probe and DNA is higher in comparison with the stability of the mismatch. The hairpin makes the mismatch hybrid thermally less stable than the exact match. Therefore, the hairpin is open up only in the presence of the more stable hybrid viz, during exact base pairing.
Furthermore, the stability of the complementary base pairing is far more as compared to the stability of the stem base-pairing therefore once the exact match is found the stem unfolds.
Designing the molecular beacons:
Designing the molecular beacons are quite a tedious job but it will give a good output. There are several points you have to understand if you want to design your own beacon.
Designing the primer:
The primer used for the real-time quantification must follow all the standard rules of primer designing. Read our article on primer designing: PCR primer design guidelines. Broadly, the primer has the following characteristics:
-
-
- 18- 2o nucleotides long
- 50 to 55% of GC content
- No secondary structure formation
- No complementary bases
- Annealing temperature between 55°C to 65°C
-
The annealing temperature of primers and beacons must be the same or nearer.
Designing the target:
Target designing is as important as primer designing.
The amplified product of the target DNA must be between 100 to 300 base pairs. A fragment larger than this will create unnecessary problems in the PCR amplification. The target region must be less GC rich and can be free of secondary structures so that the molecular beacon can bind efficiently.
Designing the molecular beacon:
Design beacon in two different parts, first design the loop part and then the stem part. The loop must have a controlled GC content and melting temperature 7 to 9°C higher than the annealing temperature.
The annealing temperature of the beacon must match the annealing temperature of the primers or we can say with the target region. Put simply, at the same annealing temperature the primer and probe bind to the complementary region.
The maximum nucleotides in a loop are 30, do not use more than this. The stem contains 5 to 7 complementary bases.
Keep one thing in mind, the stability of the stem must not be more than the stability of the probe and DNA hybrid.
If the stem base pairing is more stable; then the loop nucleotides cannot bind to its complementary sequence.
Nonetheless, the stem base-pairing must be stable enough that it can not unfold in the presence of mismatch base pairing of the loop. Check the melting temperature of the molecular beacon using the melting curve analysis.
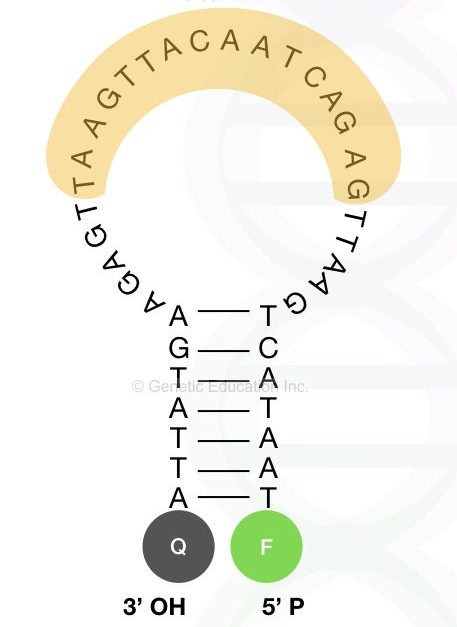
Adding the fluorochrome and quencher dye:
A single fluorochrome added at the 5′-OH end of the beacon while the quencher molecule is added to the 3′ -P end in close proximity of the fluorochrome.
Advantages of molecular beacons:
- It increases the specificity and the sensitivity of the reaction.
- It increases the thermal stability and efficiency of hybridization.
- Probes can be preserved during the reaction.
- The molecular beacons facilitate the detection of multiple targets using multiple fluorochromes in a single reaction, efficiently.
Applications of molecular beacons:
In comparison to the dye-based method, beacon chemistry is more valuable in the diagnostic industry as it facilitates higher specificity.
It is used in the determination or separation of different alleles.
In microbial genetics, the present technique is employed in order to detect and measure pathogens from various samples.
Multiplexing is easier using the molecular beacon probe-based method. By using the differently fluoro-labeled molecular beacons, different alleles or templates in a sample can be amplified and quantified.
Using the beacons in the endpoint real-time PCR techniques do genotyping precisely.
The present probes also have a significantly important role in gene expression studies, gene quantification and studying copy number variations.
SNPs (single nucleotide polymorphism) studies also use the present probe-based chemistry.
Probes are also used in viral load quantification.
Interestingly, it is employed as an intercellular probe for the detection of DNA or mRNA.
Read more: Inverse PCR: Principle, Procedure, Protocol and Applications.
Conclusion:
The molecular beacons provide the next level of sensitivity in diagnostic practices. Ready-to-use molecular beacon kits for different templates are available nowadays. The probe-based method is more accurately quantifies the nucleic acid than the dye-based method.
Conclusively, we can say, the molecular beacon probes aren’t used commonly in every RT-PCR reaction. Instead, only used when a higher specificity is required.


