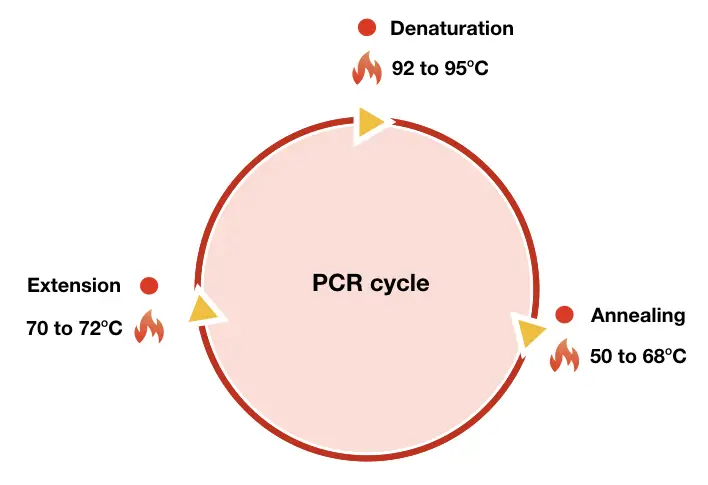
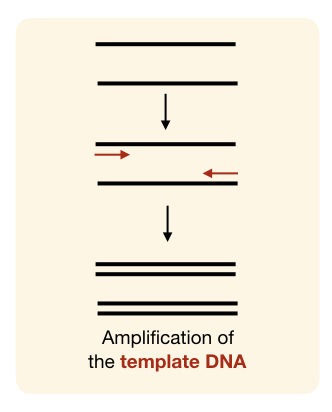
“PCR amplification is an enzyme-dependent process to copy a DNA sequence, DNA fragment, genomic DNA, plasmid DNA or any nucleic acid.“
In other words, we can say,
Producing multiple copies of DNA (in vitro or in vivo) by the enzymatic catalytic reaction is known as DNA amplification.
Amplifying DNA has crucial importance in genetic research. Nearly every experiment (in genetics) relies on PCR amplification. The reason is pretty simple,
We need tons and tons of DNA to study. A specific gene or DNA sequence can’t be studied by only a lot of extracted DNA. So we require copies/lots of copies of that particular gene.
And that’s what PCR amplification is!
The idea of PCR amplification was originally derived from cells’ natural replication system. A cell’s replication system synthesizes new DNA from the parental strand and transmits it to newly formed daughter cells.
The catalytic activity of enzyme DNA polymerase assists the synthesis of new DNA in a cell. Unfortunately, unlike this, we need a thermostable DNA polymerase to mimic the process. A Taq DNA polymerase is one among those thermostable DNA polymerases that have the power to amplify the DNA.
Amplification is a simple process but requires expertise and experience to achieve excellent results. A good amplification provides ease and efficiency during downstream processing and interpreting results.
Unfortunately, getting a decent amplification pattern isn’t that easy. In this article, I will explain several optimization tricks or strategies to improve the quality of PCR amplification.
Read more: Polymerase chain reaction.
Key Topics:
10 Strategies to Achieve Excellent PCR Amplification
Optimize DNA extraction protocol
When you don’t get amplification, you would think you missed something to add in the reaction and that’s normal for a newbie. But that wasn’t your fault, indeed.
If the starting material, extracted DNA or genomic DNA hasn’t good quality; it can’t get amplified. Impurities such as traces of chemicals or other cell debris inhibit the process.
For example, the EDTA used during the extraction, if not removed, reduces the catalytic activity of Taq DNA polymerase and ultimately diminishes amplification rate, efficiency and quality. The purity and yield of DNA have an important role during amplification. Select DNA with a 260/280 ratio of 1.8.
Find the exact primer concentration for the reaction
Primers are an important ingredient of the PCR reaction- the process for PCR amplification. Notedly, primer concentration has a crucial impact on final results. The ideal primer concentration is 10 pM per reaction.
Sometimes, the ideal concentration isn’t enough to get good results, especially for abnormal template DNA. Impure DNA, longer target sequence or high GC content sequences are considered as abnormal templates.
When we strategically increase or decrease the concentration of primers, we can increase the chances of good results. For example, use 10 pM, 20 pM, 30 pM primer concentration and observe the results.
Select the concentration which gives you good results. Keep in mind as the concentration of primer increases the chances of getting non-specific results also increases.
Related article: How to Design PCR Primers.
Spot the type of DNA polymerase required and its concentration
Taq DNA polymerase is as important as the rest of the PCR ingredients, it’s an enzyme that synthesizes DNA at a higher temperature. Importantly, many factors govern the activity of Taq DNA polymerase which sometimes reflects in final results.
For instance, EDTA impurities decrease amplification efficiency while MgCl2 greatly increases the amplification efficiency.
In either case, you may get no or less amplification and non-specific amplification, respectively. So choosing and using Taq as per the exactly required parameter is important.
When you get no amplification or too much amplification, you can strategies Taq DNA polymerase much like the primers. You can change
- The concentration of MgCl2,
- The concentration of Taq DNA polymerase units,
- The reaction preparation time
Etc.
You also can use different polymerases like the Vent or hot-start DNA polymerase depending upon your assay requirement.
Related article: Function of Taq DNA Polymerase in PCR.
Use Gradients of Annealing temperature:
The annealing temperature is the temperature at which the primer binds with the target DNA at its complementary location. It usually ranges between 52 to 65ºC (as per my knowledge).
Once again too low and too high annealing temperatures adversely influence the final results. Meaning, give non-specific binding and no amplification, respectively.
Adjusting the annealing temperature has been a crucial strategy in PCR technology and that works too! However, primer designing software advises the annealing temperature for the primer set we design.
Students wonder that it’s 100% correct and accurate! But we can get 10 fold more good results by adjusting the Ta (annealing temperature). It’s a simple process.
Prepare the same type of reaction for different types of the temperature gradient. For example, if your advised Ta is 58ºC; you can prepare gradients of 56ºC, 57ºC, 58ºC, 59ºC and 60ºC.
The strategy not only gives you efficient results but also helps you to measure the probabilities of non-specific bindings and lack of amplification.
Related article: Optimize your PCR reaction using the Gradient PCR.
Control the use of PCR additives:
PCR additives such as MgCl2, KCl, DMSO and BSA, etc have great value to enhance PCR amplification. However, they do not have a direct effect on amplification. It is also important to note that adding PCR additives unnecessary will mix up and mislead results.
For example, when we add MgCl2 unnecessarily, it will produce many DNA bands and give non-specific results. It will even produce primer dimers.
So PCR additives are used if only required during the assay. When you will notice less amplification, unclear banding pattern, primer dimers, more DNA bands and faded banding pattern, you can surely look for the appropriate additive.
Identify which additive you need, and strategies the gradient experiment to examine its exact effect on final results.
Related article: What are the different components used in the PCR reaction buffer?
Prepare a good composition of PCR master mix:
The PCR master mix is a mixture of various PCR ingredients such as dNTPs, buffer, and additives required for amplification. Ideally, ready-to-use PCR mastermixes are preferred by research students.
But when we are talking about critical experiments and diagnostic samples, it’s advisable to make our own, as per our experimental requirements. For example, a 200 µM concentration isn’t sufficient for long-range PCR.
However, it also requires optimization to get the right combination of chemicals but when you get your own combination, you can use it for repeat experiments.
I strongly recommend using separate ingredients and use as per the requirement of the assay.
Related article: 10 secrets to Prepare a PCR reaction.
Use a different PCR cycling condition:
When you set more cycles you will get more amplification and vice versa. But that’s not the case. There is a threshold above which the machine unnecessarily runs but doesn’t give amplification.
We can’t find that threshold level as it depends on the concentration of PCR reagents, template and primer used. For example, when all the primers are utilized, the reaction can’t amplify more DNA.
Ideally, 30 to 35 cycles are recommended but you can get excellent amplification results with lower cycles as well. This saves time, energy and resources.
So I advise using gradients of different cycling conditions to choose the appropriate number of cycles.
Related article: Optimizing PCR Cycles to Get Good Results.
Adjust the protocol to amplify longer templates:
It’s scientifically proven that we need a different experimental setup and reaction conditions to amplify longer templates.
Put simply, these types of templates need more ingredients, longer cycling conditions and a different type of Taq DNA polymerase. You can read this article to learn more:
Different setup for GC rich templates:
Much like the longer DNA templates, GC-rich templates are hard to amplify as well. Scientists use different methods and buffer systems for GC-rich DNA but not all work all the time.
I strongly advise using a ready-to-use GC-rich buffer which gives excellent results most of the time but that is also not enough sometimes. In those cases, you need to rethink your target sequence, primer sequence, primer location and complementation.
If possible, choose some other region of the template that is less complex and has less GC content, try to lower down the GC content as much as possible.
Ideally, the GC content must be between 45 to 55%.
Related article: What is the importance of GC Content?
Wrapping up:
These strategies are working but you have to perform many optimization experiments and that is indeed needed in genetic research and diagnostic labs. Once you have a proven and optimized protocol, recipe and reaction conditions you can replicate it for years.
Especially, for COVID testing do optimization to achieve good amplification; it will ultimately benefit you in the long run. I hope this article will add more value to your PCR knowledge.
External resources:
- Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol. 2013;133(3):1-4. doi:10.1038/jid.2013.1. Article link: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4102308/.