Capillary electrophoresis abbreviated as CE is an analytic technique for measuring DNA separation based on its size, charge and separation time. It’s high throughput, accurate and faster separation technique.
Electrophoresis has been significantly important in analytical biology, especially for investigating DNA. The usual two common types are native agarose gel electrophoresis and polyacrylamide gel electrophoresis.
Put simply, native electrophoresis is applied for routine investigations and PCR amplicon analysis while PAGE is applied to separate smaller fragments and is capable of separating DNA fragments during sequencing.
The principle of the electrophoresis technique explains that particles can be separated based on their electrical charge, size and medium used.
Although either technique has important consideration in genetic research, they come up with serious shortcomings. The electrophoresis technique is slow, time-consuming, less accurate and requires more starting sample volume.
It has been a serious problem when testing forensic samples like hair DNA or saliva traces. Hence a high-throughput, faster and accurate separation technique was needed since long and ultimately the problem was solved by capillary electrophoresis.
But what exactly is capillary electrophoresis! Is it useful? And when it’s required?
In the present article, I will explain the concept of capillary electrophoresis, principle, instrumental setup, applications and advantages. I will also explain the protocol and how it works for DNA especially.
Stay tuned.
Key Topics:
What is Capillary gel electrophoresis:
Capillary- a submillimeter hollow tube
Electrophoresis- a separation technique
Read more: What is Gel Electrophoresis and What is it used for?- A beginner’s Guide.
The capillary electrophoresis abbreviated as CE, consists of nonfluid channels and submillimeter capillaries which separate molecules under high electric current. Contrary to the micro separation PAGE, it’s a nano-level, particle separation technique.
Anes Tiselius (1930), first explained the concept of CE, however, Jorgenson JW & Lukacs KD described the technique precisely.
In a layman language, the basic function the CE performs is to separate particles (any nanoparticles but here we will discuss the DNA only). You may wonder why CE and not other native electrophoresis?
See, the native electrophoresis technique is a manual technique and isn’t a high throughput one! It can’t separate minute particles or DNA fragments. For instance we can visualize 100bp, 200bp or 500bp fragments but can’t investigate 100bp, 101bp or 109bp fragments.
That’s nearly impossible.
Besides, the native electrophoresis technique is
- Tedious and time-consuming
- Less effective
- Costlier
- Hazardous
- Limited
- Inaccurate
Moreover, It can’t determine small sequence variations which unfortunately restricts the analysis strength. Fortunately, the CE can solve almost all problems enlisted here.
Principle of capillary electrophoresis:
When a sample is injected using pressure or electrokinetic injection into the capillary, filled with a conductive buffer fluid, charged particles separate based on mobility, size and electrical current.
Under the influence of high current (300V/cm) particles like fragments of DNA migrates from negative to positive end at where the capillary ends into the destination reservoir. Smaller DNA runs faster than the larger one, the detector located in-between detects the smaller to larger DNA, speed of separation and mobility.
The entire principle of the CE relies on two factors which are the mobility of particles and Endo-osmotic flow (EOF). Here the mobility is decided by the charge and size of the article while the EOF is decided by which and in which amount the capillary filled material is charged.
UV absorbance and fluorescence analysis are two common detection methods used for CE in which fluorescence-based detection is widely used for instruments used in DNA analysis.
“Charge to size ratio of molecules is taken into consideration for capillary electrophoresis assay.”
How is it different?
It is important to discuss and understand that the CE is different not only then the native electrophoresis technique but also different from HPLC and high-pressure gas chromatography.
The difference is its principle which relies on the “Endo-osmotic flow”
The buffer system of source and destination reservoir, cathode & anode placed, the structure of capillary and its connection with the power supply creates a strong electrical circuit that helps run the sample.
Instead of high pressure or gas, the chemical coat present in the inner wall of the capillary creates a smooth endo-osmotic flow. Take a look at the example.
To separate the negatively charged DNA, the inner capillary wall is coated with a cationic surfactant. The cations react with the anions present in the buffer, create a double charge layer on the wall and provide a way for DNA to migrate.
Take a look at the figure below,
Diagram of Capillary electrophoresis:
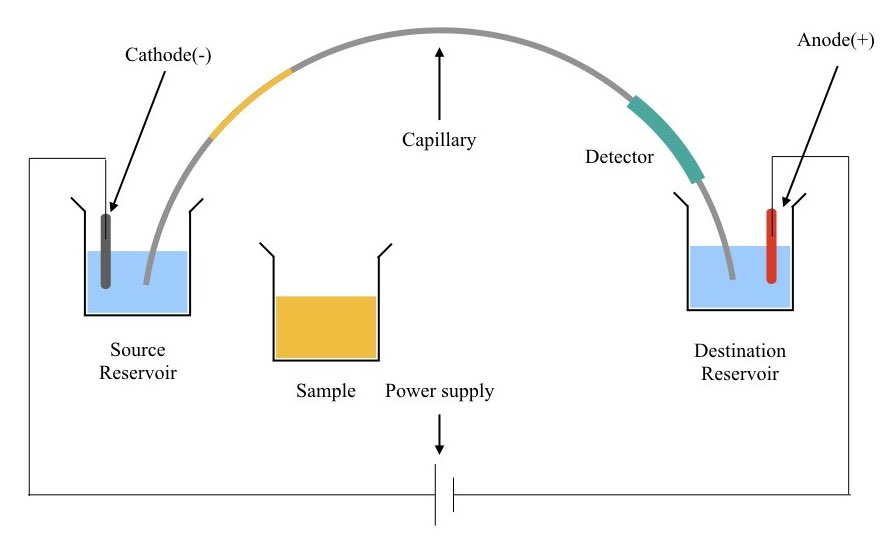
(I hope this explanation helps you to understand the principle and basic concept.)
So basically, it accurately separates every fragment as well as eliminates the requirement of high-pressure sample injection.
“Studies also suggest that the DNA has a constant charge and size ratio therefore in the free solution its electrical mobility remains the same which disallows accurate size base determination.
CE fixes this problem by allowing the sieving matrix filled capillary system which readily separates DNA particles depending upon their size, exclusively. This feature changed the entire sequencing technique as each and every DNA base can be investigated and sequenced accurately.”
We will discuss more on it in the application part.
Read further: Factor Affecting DNA Agarose Gel Electrophoresis Results.
To understand the principle more precisely let us go through the instrumental setup for the capillary electrophoresis.
Capillary electrophoresis instrument:
Components:
- Sample vial
- Source reservoir
- Destination reservoir
- Capillary tube
- Electrodes- cathode and anode
- Power supply
- Detector
- Data processing device- computer system
- Temperature controller
Source reservoir: It is filled with the aqueous buffer which provides the constant buffering environment during the run.
Destination reservoir: it is also filled with the aqueous buffer that protects the sample after completion of the run.
Sample via: The sample vial consists of the sample or samples to run in which the capillary is placed.
Capillary tube: The capillary tube is a submillimeter micro tube filled with either silica or coated with polyimides. The tube helps migrate charged particles to the destination reservoir.
Electrodes: electrodes cathode and anode are placed in each reservoir and complete the electrical circuit which facilitates the migration of charged particles from one to another end.
Power supply: The power supply provides a constant electrical current during the run usually is high-voltage current.
Temperature controller: Increment in instrumental temperature reduces the strength of viscosity during the run and creates problems during migration, so temperature controller units are provided in several instruments.
Capillaries for CE:
The capillaries used in the capillary electrophoresis of DNA, capillary DNA sequencing, capillary SSCP or forensic analysis should have several specific characteristics.
The capillary utilized here is a long hollow tube with 50 to 75 μM diameter and 3o to 60 cm long. It is coated with fused silica mostly for DNA studies. Notedly, the polyimide is used as a protecting coating outside.
However, coating inside also increases the separation of molecules hence some capillaries are also available with inner coating as well. These types of capillaries without polyimide coating are highly recommended for UV-based detection methods.
The transparency provided by the fused silica provides ease during UV-VIS detection. Moreover, it is cheaper, inert, easy to handle and doesn’t require additional inner or outer coating.
The capillary length here plays a crucial role. A shorter capillary increases the separation and cuts down the time and cost while the longer capillary may compromise separation efficiency and elevate time and cost. Although longer capillary increases resolution strength.
The micrometer size of the capillaries facilitates good heat dissipation and allows higher current effectively.
Detection, results and analysis:
Broadly, there are two common ways to detect the separation of molecules in CE which are UV-VIS analysis and fluorochrome-based analysis.
Much like the UV-VIS spectroscopic analysis a UV-light beam when passed through the sample detects DNA fragments and gives the signal. Notwithstanding, as the technique highly relies on the path length it’s less advisable.
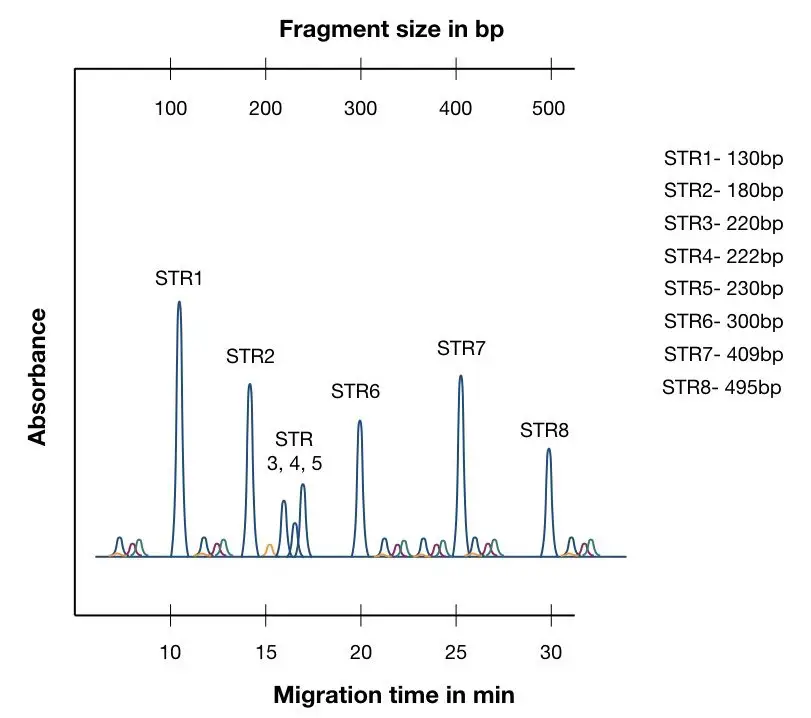
Contrary, in the fluorochrome-based analysis, the molecule or here DNA must be first coated with the fluorochrome dye in order to track their migration. When the fluorochrome emits fluorescence a detector detects signals and converts them into graphs.
Usually, it is known as an electropherogram which provides information about the fluorescence absorbed by a molecule, distance traveled and size of the fragment. It looks like this. To know more, read our article: What is Electropherogram? How to Read it?
Advantages of Capillary electrophoresis:
The CE has evolved so fast in DNA analysis and sequencing because of its tremendous advantages over conventional techniques.
No gel preparation step is required:
It’s safer than conventional gel electrophoresis as it involves the use of harmful chemicals like agarose or EtBr.
Faster than usual:
One of the important advantages the technique provides is the speed of separation. The present technique is much faster than other conventional techniques. Conventional gel electrophoresis requires 1 to 2 hours from preparing a gel to running the samples. CE can do its job in minutes.
Reusability at its height:
One of the important advantages the EC has is the reuse of capillaries. It can be reused after washing and gives excellent results, though.
High throughput:
To automate most of the applications in genetics, the technique should be high throughput, meaning it can handle multiple samples at a time without compromising the accuracy and speed.
CE is one of the most innovative high throughput genetic techniques which allows the use of multiple capillaries to run multiple samples. 12 samples of 96 sample capillary setups are now available.
High resolution and precision:
As mentioned before, the technique has nano-level separation capability and therefore it can separate even a single base pair precisely. No other separation technique can do this job as effectively, faster and accurately as the CE.
Complete automated system:
Conventional electrophoresis is a total manual method, needs time, manpower and is also prone to human errors. CE is a fully automated separation technique that does not even run samples automatically but generates reads and evaluates results automatically.
Low sample consumption:
The idea behind developing the CE has been to reduce the sample consumption and sample preparation step. The low sample consumption makes it readily available for forensic sample analysis and DNA sequencing.
Fascinatingly the sample required for the single run is as low as 1 (nL) nanoliter.
Accurate sample handling and automated sample loading:
Yet another crucial benefit the technique provides is sample automation. DNA samples can be loaded automatically and pre-preparation steps aren’t required either.
Techniques like GC or HPLC seek additional utility for sample injection which isn’t required in the capillary electrophoresis too.
Applications of Capillary electrophoresis:
As discussed previously, here we are only discussing applications of the capillary electrophoresis technique for DNA analysis or in genetics only. It is applied in
- Forensic analysis and STR typing
- Genotyping and native DNA analysis
- Denatured DNA analysis
- Investigating SSCP (SIngle-strand conformation polymorphism)
- Capillary DNA sequencing
Capillary electrophoresis in Sanger sequencing:
Currently, the CE is used popularly in Sanger sequencing. In the process, it provides automation, speed and accuracy. Basically, the principle of the Sanger relies on the termination of the chain.
Here instead of dNTP, a ddNTP binds with the growing DNA chain and terminates its amplification. The single base pair separation resolution of CE provides a tremendous advantage in this technique.
A fragment of every single terminated sequence of even a single base pair can be accurately and precisely separated and examined.
And therefore the CE has great value in the sequencing platforms. It is also used in other sequencing platforms too.
Related article: DNA Sequencing: History, Steps, Methods, Applications and Limitations.
Forensic analysis and STR typing:
Yet another popular and widely accepted application of CE is in STR typing and forensic analysis. STR stands for Short Tandem Repeats, meaning short repetitive sequences, located one after another.
The number of repeats varies among individuals and therefore STR marker is widely used. However, shorter fragments are hard to analyze on a gel.
CE here separates every single fragment based on its repeat numbers. Hence it facilitates accurate identification of biological samples. Forensic departments across the world trust the capillary technique only.
Related article: DNA Fingerprinting- Definition, Steps, Methods and Applications.
Wrapping up:
Capillary electrophoresis is one of those techniques that genetic analysis needs nowadays. In this COVID condition, CE facilitates speed and trustworthy outcomes. However, these techniques aren’t yet that much automated.
When combined with sequencing, it increases the overall cost, in comparison, with the conventional sanger sequencing.
In my personal experience, though CE is tremendously advantageous still smaller genetic setups can’t afford it. I think cheaper and faster options are required, especially for whole-genome sequencing.
Resources:
Durney BC, Crihfield CL, Holland LA. Capillary electrophoresis applied to DNA: determining and harnessing sequence and structure to advance bioanalyses (2009-2014). Anal Bioanal Chem. 2015;407(23):6923-6938. doi:10.1007/s00216-015-8703-5.

