“miRNA is an endogenous dsRNA that participates in gene regulation. Explore this complete guide to learn the miRNA-mediated gene silencing in this article.”
Fun Facts:
The human genome may encode 1900 different miRNAs.
Over 60% of human coding genes are regulated by the miRNA-mediated pathways.
40% of miRNAs are located in the intronic regions.
As we discussed, some of the miRNAs are highly conserved in all mammals; these miRNAs have, on average, more than 400 different conserved target mRNAs.
Further, around 16% of pre-miRNA is affected by the RNA editing process (which can not work properly during gene silencing).
Chronic lymphocytic leukemia is associated with the dysregulation of miRNAs.
In 2024, the Nobel Prize in Physiology or Medicine was awarded to Ambros V and Ruvkun G for their groundbreaking contributions to microRNA and exploration of its role in gene regulation.
And the story of non-coding RNAs, yet another mysterious element of our genome, sparked again! Although the fundamentals were established in 1993, these scientists continued the previous work into molecular medicine applications.
miRNAs are different from mRNA. They are non-coding! But what makes them interesting to study is their role in gene regulation. They bind with mRNA and control gene expression.
Want to learn how it happens?
Check out this article to learn about microRNA, their biological significance, function, types, processing, degradation, applications and a lot more.
Stay tuned.
| Abbreviation | Full name |
| mRNA | Messenger RNA |
| miRNA | microRNA |
| shRNA | Small hairpin RNA |
| siRNA | Small interfering RNA |
| RNAi | RNA interference |
| pri-miRNA | Primary microRNA |
| pre-miRNA | Precursor microRNA |
| RISC | RNA-induced silencing complex |
| Ago | Argonaute protein |
Key Topics:
What is miRNA?
The miRNA, short for microRNA, are short, double-stranded, 22 to 25 bp in size and natural RNA molecules present in eukaryotes. Some miRNAs are also found in viruses, though.
They have been present in our genome for millions of years in the form of conserved sequence domains. Meaning, during evolution, these sequences remained unchanged and unaffected by natural forces.
This shows how important their function is, even though they don’t participate in protein synthesis. The first microRNA was identified in 1993 by a group of scientists led by Lee and Ambros.
We will discuss the detailed history later in this article.
The number of miRNA vary among various species with the same function of gene regulation. Check out this table for miRNA in different organisms.
| Organism | Number of miRNAs |
| Human | 695 |
| Chicken | 475 |
| C. elegan | 154 |
| Zebrafish | 337 |
| D. Melanogaster | 152 |
| A. thaliana | 187 |
If you want to know more about the number of miRNAs, you can check out the database named miRNABase.
History:
In 1993, while studying post-embryonic development in the model organism C. elegans, the team of scientists led by Lee and Ambros discovered the microRNA. They believed that it’s only present in nematodes.
What they had observed was that the lin-4 gene could be transcribed into the mRNA but not translated into the protein. Further research showed that the lin-4 transcribes into a 20 to 60 bp short RNA.
Interestingly, this short and double-stranded RNA participates in the lin-14 gene regulation by an antisense mechanism.
Later on in 1998, Fire A and Mello C published a paper explaining an interesting mechanism, referred to as RNA interference. They deciphered double-stranded RNA-mediated gene expression regulation.
In 2000, a group of scientists led by Ruvkun G. discovered the second microRNA let-7 from C. elegans. Further studies showed that the miRNAs are conserved across species.
These findings laid the foundation of a universal mechanism regulating gene expression mediated by microRNA.
After 2001, various miRNAs have been identified from different species, including plants, humans and other animals. The miRNABase was established after that. It comprises the database and information on the miRNA.
Afterward, Ambros V and Ruvkun G were awarded the Nobel Prize for their contribution to the field of microRNA.
Structure of miRNA:
As we stated, the microRNAs are shorter, double-stranded RNAs, but let me tell you, they aren’t true double-stranded, initially!
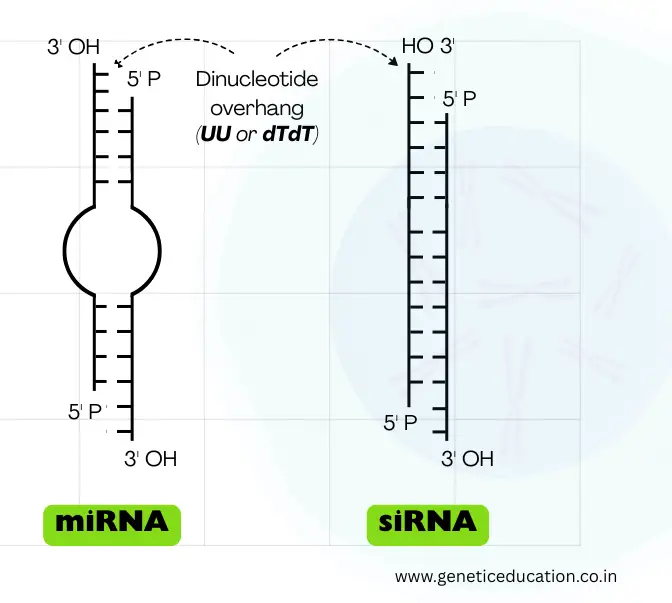
It folds into the hairpin (stem-loop) structure and becomes double-stranded. However, like the mRNA, it has a 5’ cap and 3’ poly-A tail, also. Note that its structure changes with each step of processing.
The stem consists of ~33 to 35 base pairs, while the loop contains roughly 10 to 30 nucleotides. The loop is variable in length. It also consists of the classic characteristic of the double-stranded RNA— a dinucleotide overhang at the 3’ end.
Now, this structure usually appears during the pre-miRNA stage; upon maturation, it becomes a 22 to 60-nucleotide-long double-stranded RNA. In the double-strand, one strand is known as the guide strand while the other strand is known as the passenger strand.
Location of miRNA:
Research suggests that even though the miRNA domains are highly conserved, their location within the eukaryotic genome varies. They are located randomly either in intergenic regions, introns or even exons.
- The majority of miRNAs are located in intergenic regions— between two genes.
- Within the introns of a gene.
- Within the exons of a gene.
Based on that, they either transcribe independently using a dedicated promoter, co-transcribe with a gene using a shared promoter, or transcribe as a single unit or shared unit.
In addition, they are also present as an independent single unit (monocistronic) or in a group or cluster of many miRNAs (polycistronic).
Nomenclature:
The nomenclature was originally developed and refined by Bernardo et al. and is managed centrally by miRBase, the official database of microRNA sequences. The miRNA nomenclature is based on these factors:
- Species origin
- Maturation state (precursor vs. mature)
- Strand of origin (5p vs. 3p)
- Gene origin (if transcribed from multiple loci)
- Functional role (guide vs. passenger strand)
The nomenclature starts with a species prefix, for example– hs-miR-21: human miRNA 21. Some common prefixes are listed here:
- hsa = Homo sapiens (human)
- mmu = Mus musculus (mouse)
- rno = Rattus norvegicus (rat)
- ath = Arabidopsis thaliana (plant)
In addition, ‘mir’ is used for precursor miRNA, while ‘miR’ is used for mature miRNA. For example,
→ hsa-mir-21 = Human precursor miRNA 21.
→ hsa-miR-21 = Human miRNA 21.
The 5p is used for the 5′ arm and the 3p is used for the 3’ end. For example,
→ hsa-miR-142-5p = mature miRNA from the 5′ end
→ hsa-miR-142-3p = mature miRNA from the 3′ end
Previously * has been used for the 5p/3p system.
→ miR* for the passenger strand
→ miR for the guide strand
For example, as per the older system: hsa-miR-20a (guide), hsa-miR-20a* (passenger).
As per the modern system: hsa-miR-20a-5p (from 5′ arm), hsa-miR-20a-3p (from 3′ arm).
Function:
The miRNA doesn’t participate in the protein synthesis process, instead, it binds with the mRNA and guides it for degradation or suppression. By doing so, it regulates gene expression.
Besides their primary function of gene regulation, they also have other important functions. For instance—
Regulation of metabolic pathways– lipid metabolism, immune system, cell proliferation, apoptosis, neuron patterning, hematopoietic stem cell differentiation, etc.
During the RNA interference, their dinucleotide overhang also helps to choose between the guide and passenger strand, recognized by the argonaute protein and processed by RISC (RNA-Induced Silencing Complex).
In plants, they go one step further and participate in leaf and flower development. They control all these activities at an epigenetic level.
Some of the eukaryotic (human) miRNAs and their functions are explained in the table below.
| miRNA | Gene target | Function |
| miR-16 | Any gene | AU- rich elements mediated mRNA instability |
| miR-146 | c-Myc, ROCK1 | Development of the immune system |
| miR-17-92 | c-Myc, E2F1 | B-cell lymphoma upregulation |
| miR-143, 145 | Unknown | Downregulation of colonic adenocarcinoma |
| miR-32 | PFV1 (retrovirus) | Antiviral defense (cleaves exogenous dsRNA) |
| miR-143 | ERK5 | Differentiation of adipocytes |
How miRNA Regulates Gene Expression?
In this section, we will deep dive into the mRNA processing part and understand how it regulates gene expression.
This long journey of miRNA-mediated interference is divided into three broader steps—
- Processing in the cell nucleus
- Processing in the cell cytoplasm
- Processing for degradation
Note that it follows a similar processing pattern to the small interfering RNA and shRNA with minor modifications.
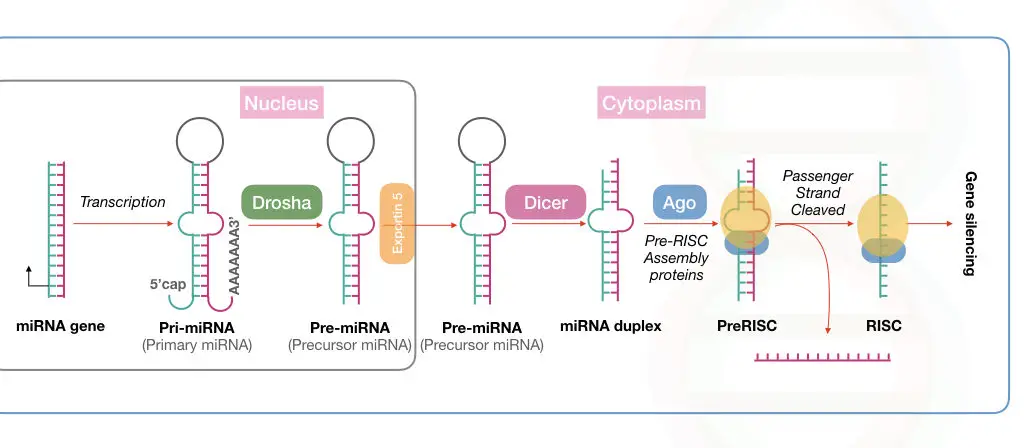
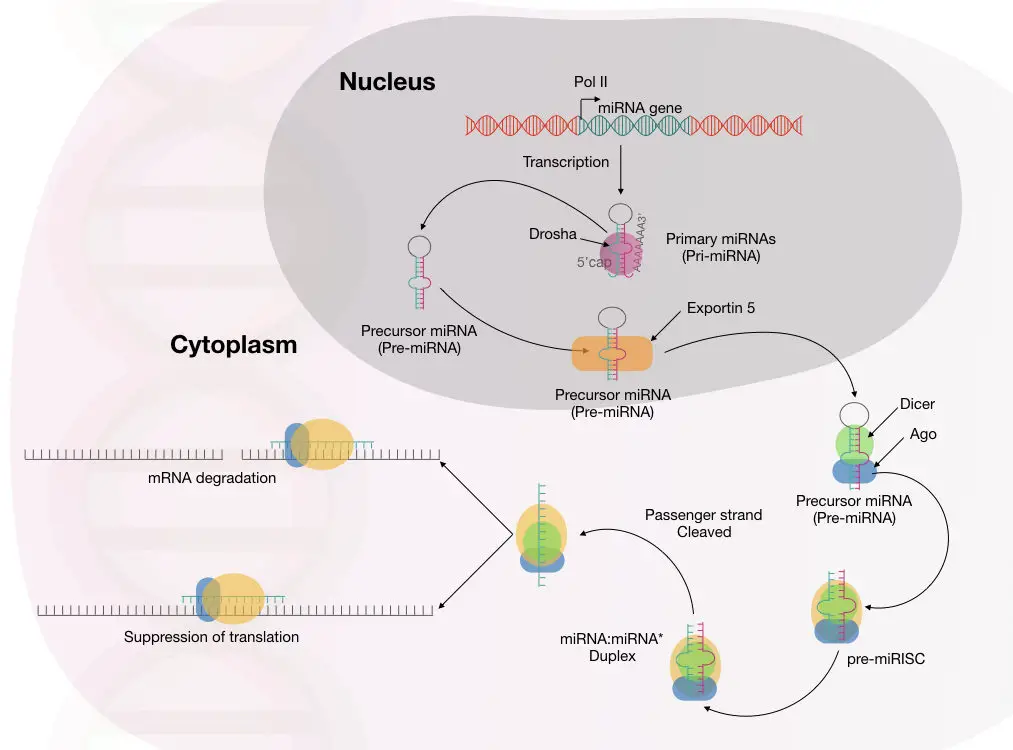
miRNA processing in the nucleus:
The processing begins with the synthesis of pre-miRNA transcript by the transcription. This happens in the cell nucleus.
RNA polymerase II and III participate in this process, though RNA polymerase II is involved in the transcription of the majority of miRNAs.
Now, the transcribed products from RNA polymerase are known as the primary microRNAs or pri-miRNA, in short. Note that this primary transcript is several kilo base pairs in size with a hairpin or stem-loop-like structure.
This stem-loop structure must be cleaved off for mature miRNA synthesis via precursor miRNA. Drosha, a type of RNase III enzyme, forms a large complex to initiate the precursor formation by digestion.
The large microprocessor complex, formed by Drosha and DGCR8 or ‘Pash’, performs this task effectively. Till now, the mature miRNA has not been created.
This cleaved structure is known as the precursor miRNA (shown in the image) formed from the activity of the microprocessor complex.
This Drosha-mediated catalytic activity forms the double-stranded miRNA hairpin duplex structure, generating the 5’ phosphate and 3’ OH with dinucleotide overhangs.
Now, our raw material for miRNA maturation is ready! It will undergo the process of transportation to the cytoplasm for further processing.
miRNA processing in the cytoplasm:
The protein transportin 5 controls the process of miRNA transportation from the nucleus to the cytoplasm. EXP5 or exportin 5 is a RanGTP-dependent nuclear transport receptor protein.
Here, RNase III, also referred to as Dicer, forms the mature miRNA from the precursor miRNA. A highly conserved protein, Dicer, is a specific type of nuclease that cleaves the pre-miRNA exactly at 22 nucleotides and generates a mature miRNA duplex.
Notedly, in plants, the miRNA maturation process occurs in only the nucleus.
Immediately after that, the mature miRNA is loaded onto the Argonaute protein. By interacting with other important proteins, the miRNA is loaded into the RISC, where the passenger strand is removed and the guide-strand is selected to use as a complementary strand against the target mRNA.
RISC:
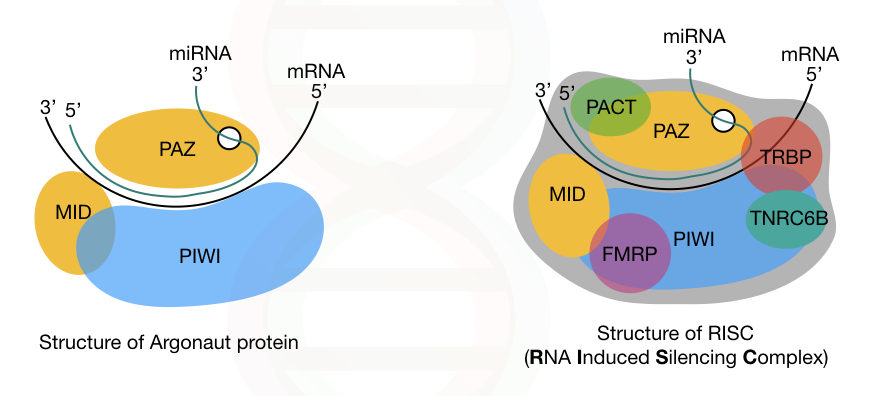
Now let’s talk about the important complex in the RNA interference process— the RISC.
RISC is an RNA-Induced Silencing Complex. It is a type of ribonucleotide protein complex that includes various proteins, besides the miRNA guide-strand. Whereas, Dicer, argonaute, and other primarily protein, are the major constituents of RISC, other minor proteins are,
- TRBP
- PACT
- TNRC6B (RNA recognition motif-containing protein)
- FMRP (fragile X mental retardation protein)
- Tudor staphylococcal nuclease domain-containing protein and putative DNA helicase MOV10.
In addition, three different domains of the key protein, argonaute, are PAZ, PIWI and MID. The PAZ domain binds to the 3’ end of the miRNA-guide-strand while the PIWI domain interacts with the 5’ end of the miRNA.
Note that the PIWI domain is structurally similar to the ribonuclease-H; however, it has a different function. Our genome consists of genes for eight different argonaute proteins.
The complete structure of Argonaute and the RISC is explained in the image below.
In the last step, the RISC undergoes the gene silencing process using either of these two mechanisms: Translational repression or mRNA degradation.
Studies showed that multiple miRNA binding sites are present on the 3’ untranslated region of the mRNA.
Among the two mechanisms, mRNA degradation is widely studied and well-accepted. Here, the miRNA-guide-strand binds with the target mRNA near the 3’ untranslated region.
To process the mRNA using this mechanism, the miRNA guide-strand must be highly complementary to the mRNA. Immediately after this, the Ago protein complex slices the mRNA and destroys it.
The mRNA exonuclease digestion is followed by two different events: 5’ end decapping and deadenylation.
In another mechanism, if the miRNA guide strand isn’t complementary to the mRNA, it still remains attached to the mRNA using a few nucleotide matches and blocks the mRNA processing further.
This usually represses the mRNA and does not cleave it. The complex mechanism of mRNA repression using this mechanism is not clearly understood.
In addition, multiple other mechanisms for gene silencing are listed here.
- Binding to the 5′ UTR instead of the 3′ UTR to regulate gene expression.
- Regulation of other small RNA species or other miRNAs by a specific miRNA.
- Induction of poly-A tail shortening, leading to repression of mRNA translation.
- Inhibition of efficient translation by the ribosome, reducing protein synthesis.
So, it is now evident, scientists have reported multiple mechanisms for gene silencing mediated by miRNA. The complete molecular mechanism of miRNA-mediated gene silencing is explained in the image below.
Interestingly, the process of miRNA maturation is slightly different in plants. Some differences are listed here.
- The plant miRNAs often have a 5’ cap and a poly-A tail, like the mature RNA.
- The plant miRNAs have a slightly larger hairpin loop.
- They also contain the miRNA isoforms.
- The maturation of miRNA occurs only in the nucleus; instead of Dicer, the dicer-like 1 protein, called DCL-1, mediates the miRNA maturation.
- Instead of Exportin, the HASTY (HST) mediates the transportation of mature miRNA from the nucleus to the cytoplasm.
- However, the miRNA loading in the Ago protein of RISC occurs either in the nucleus or the cytoplasm.
- In plants, the miRNAs are not clustered and thus not transcribed from a single promoter; each miRNA has its own transcription machinery.
miRNA degradation:
Now, our miRNA enters its final life stage— the degradation!
miRNA degradation is required upon completion of its function. The reason is, they can target multiple mRNA and process them for degradation. So to avoid unwanted gene silence, it’s crucial to destroy the miRNA.
Smaller RNA-degrading nucleases, ribonucleases, exonucleases, and endonucleases with their 5’ to 3’ and 3’ to 5’ activities, cleave and degrade the miRNA.
miRNA dysregulation:
This is the complete journey of the miRNA and its role in the gene silencing process. However, aberrant miRNA mechanisms cause serious health-related issues. It results in loss-of-function or gain-of-function mutations.
Studies showed that overactivity of miRNA leads to downregulation of genes, while the underactivity of miRNA leads to upregulation of genes and causes abnormalities.
miRNA abnormalities can result in obesity, some nervous system diseases, kidney disease, heart disease, cardiovascular disease and cancer.
miRNA therapies:
miRNA therapies are nowadays popular! The natural function of an miRNA can be restored in a particular tissue. This eventually helps in cancer-like conditions.
In addition, synthetic miRNAs can be designed and injected into target tissues to restore the normal function.
Scientists are also planning to regulate the overexpression of oncogenes by injecting additional miRNAs into affected tissues in cancer.
miRNA therapies hold a promising future and will revolutionize the field of cancer screening and treatment in the near future.
Applications:
miRNA has lucrative applications in various fields and scientists are trying to utilize them for healthcare. Here are some good applications.
Gene regulation:
miRNA provided a path for gene regulation research. Scientists can now better understand the post-transcriptional gene silencing, thereby, development, differentiation and homeostasis.
Disease Biomarker:
miRNAs can be used as an important biomarker, not only in cancer but also in cardiovascular and neurological disease and infection.
Scientists study the miRNA pattern during the disease and better understand the condition. This will help in disease risk assessment.
Therapeutic applications:
Synthetic miRNA will change the future of therapeutics. Such artificial molecules can restore the normal function of downregulated tumor suppressor genes and fight against cancer.
In addition, novel oncogene-specific miRNAs can be introduced to control the oncogene activities and tumor progression.
Functional genomics:
Besides these major applications, miRNA can also be employed for loss-of-function or gain-of-function mutations. Thereby, scientists can understand cell cycle regulation, immune response and apoptosis.
In conclusion, miRNA will revolutionize the oncology and cancer field in the near future. Hopefully, such therapies will help overcome cancer.
Related article: siRNA vs miRNA– Similaries and Differences.
Wrapping up:
MicroRNA plays a significant role in gene regulation. Its better understanding will, one day, help scientists decode various diseases. These shorter, double-stranded RNAs can be mimicked and used to restore the normal gene function.
However, delivering the synthetic miRNA or siRNA is still a challenge for scientists. Still, several therapies are under clinical trial and FDA approval stages. I hope you like this article. Do subscribe to Genetic Education.
Resources:
- Lee. RC., Feinaum. RL. and Ambros. V. “ The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14.” Cell. 1993; 5(75): 843-854. https://doi.org/10.1016/0092-8674(93)90529-Y.
- Wahid F., Shehzad A., Khan T. and Kim YY. “MicroRNAs: Synthesis, mechanism, function and recent clinical trials.” Molecular Cell Research. 2010;11(1803):1231-1243. https://doi.org/10.1016/j.bbamcr.2010.06.013.
- Introduction to miRNA in knowledge base by abm.
- Giza DE, Vasilescu C, Calin GA. Key principles of miRNA involvement in human diseases. Discoveries (Craiova). 2014;2(4):e34. doi:10.15190/d.2014.26.