“DNA hybridization is a molecular genetic technique in which either two single-stranded DNA or DNA and RNA are hybridized/joined by a chemical or physical mean which helps to study sequence variations.”
Scientists have been using the hybridization technique for years, let’s find out what DNA hybridization is and how it occurs.
Any genetic technology relies on either mechanism of amplification or hybridization or both. For example, conventional PCR is an amplification-based technique, FISH is hybridization-based whereas qPCR is a hybridization and amplification-based technique.
Hybridization has crucial implications in various fields including diagnosis, research and phylogenetics. It uses the similarities of DNA sequences as a foundation to compare and contrast sequences and thereby establish the relationship between organisms.
Hybridization-based approaches are conventional and traditional and have flaws, still are used in futuristic genetics as they are more flexible, robust and easy to set up. In addition, scientists can screen and identify thousands of sequence variations.
Thus, such techniques are more valuable. But what exactly DNA hybridization is, and what is the principle and applications? Let’s find out. The learning objectives of the present article are useful for students to understand the basic foundation of FISH or microarray.
Stay tuned.
Key Topics:
What is DNA hybridization?
Let’s start with some basics so that we can concrete the base.
DNA, in its natural form, is double-stranded. Two strands are joined by hydrogen bonds, two between A and T and three between G and C. Such base-pairing makes DNA duplex stable. Each strand only units if complementation occurs. Meaning A binds with T and G binds C, only.
In molecular biology, hybridization is referred to as allowing the binding of two identical or complementary nucleic acid sequences under adequate conditions. In 1960, DNA hybridization was first reported by Britten R during genome analysis.
However, the actual process of DNA-RNA hybridization was discovered by Hall B and Spiegelman S in the same year. Interestingly, the ideal and standard approach for hybridization which is still used in microarray-like assays was originally postulated by Gillespie and Spiegelman in 1965.
Traditionally the hybridization approach had a different utility. It was used for molecular phylogeny creation and establishing relations between species using sequence similarities. Let me tell you how it was done.
The completely complementary DNA duplex denatures at approximately 84°C temperature (technically the denaturing temperature is 94°C). The phenomenon is known as DNA melting. As the sequence similarities decrease, less bases share the complementation and less temperature will be required to denature them.
Separation of dsDNA duplex by heat is often known as DNA denaturation.
Scientists often mix the DNA sequences of two different species, in order to establish a relationship. In the first step, both DNA strands are denatured at a higher temperature and prevent renaturation. In the next step, both single-stranded DNA are allowed to hybridize with each other under adequate conditions and denatured once again.
Now next time, (when two single-stranded DNA of two different species are hybridized) when denaturation occurs if the two ssDNA share fewer sequence similarities, dissociation or denaturation occur at lower temperature and vice versa. Take a look at the image below.
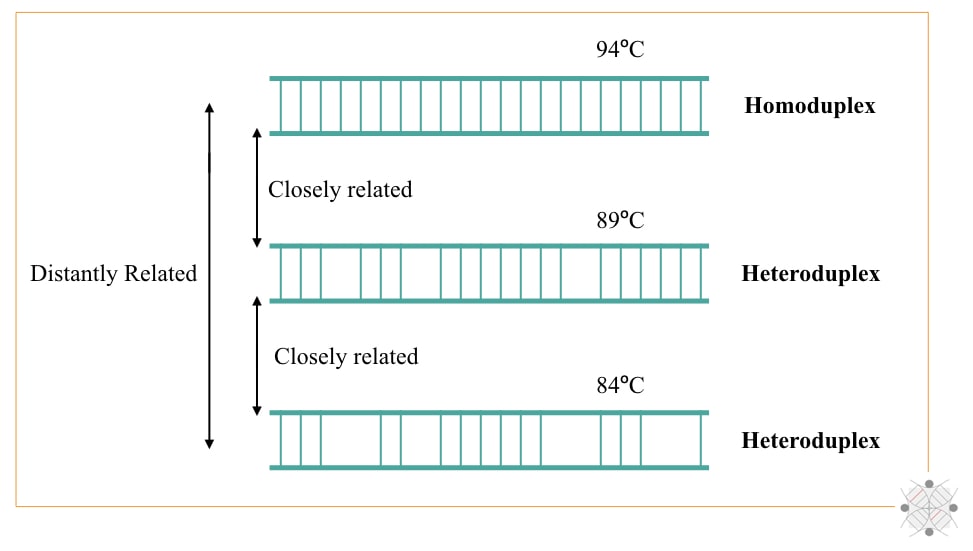
So technically, heteroduplex denatures at a lower temperature while homoduplex denatures at a higher temperature. To interpret the results, radio-labeling, UV analysis and X-ray analysis techniques were used.
Such techniques were actually employed for phylogeny analysis but are not so accurate, prone to error and less sensitive, compared to recent day PCR or sequencing-based approaches. So this was the conventional approach for utilizing DNA hybridization.
In modern times, the present technique has totally a new face and is used for finding alterations like indels, CNVs and SNPs from the genome. But before going to understand it, first, take a close look at DNA-DNA and DNA-RNA hybridization and how it occurs.
How DNA hybridization occurs?
This section explains the general principle behind DNA hybridization.
Either technique of DNA hybridization (DNA-DNA or DNA-RNA hybridization) is a type of nucleic acid hybridization completed in three steps, denaturation, hybridization or binding and washing.
The principle is stated here.
“First, the target nucleic acid or DNA sequence is denatured by heat or chemicals and allowed to hybridize with a complementary probe or another DNA or RNA sequence under adequate conditions using a hybridization buffer. In the last step, the unbound DNA template and DNA or RNA probes are washed off using a washing buffer.”
The technical approach is simply explained here.
| Step | Conditions | Outcomes |
| Denaturation | 90 to 94°C for 2 minutes | Opens dsDNA duplex |
| Hybridization | 37°C overnight. | DNA/RNA probes hybridize with the target DNA. |
| Washing | 37°C | Washing off unbound targets and DNA/RNA probes. |
Note that hybridization buffer and washing buffer are two separate chemical compositions used to boost individual processes.
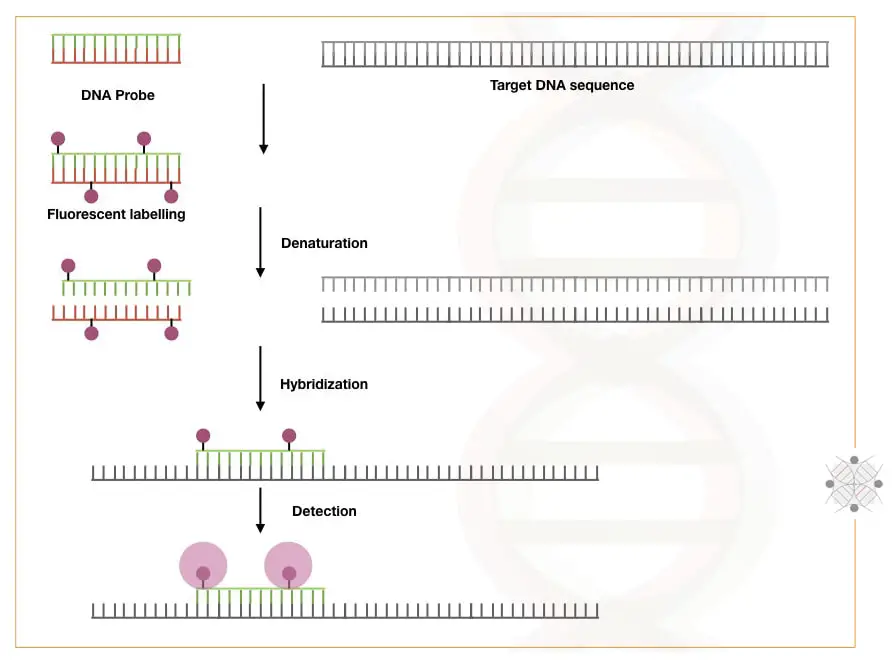
Process of Hybridization:
The technical background of DNA hybridization has three processes: Probe preparation, target or template preparation and hybridization. Let’s understand the process in steps.
Depending upon what we wish to study, or sequence variation, a probe is prepared. A probe is a short, single-stranded, known and 20 to 30 nt long nucleic acid sequence of either DNA or RNA. Probes are radio or fluoro-labeled.
The labeling process is explained in this article: DNA probe labeling.
Probes are now ready for hybridization.
For template preparation, DNA or RNA is extracted. Pre-processed, purified and fragmented for hybridization. Note that DNA is directly used for hybridization, but if you are dealing with RNA, the mRNA is first reverse transcribed into cDNA and then hybridized.
Now Denaturation is performed at 94°C followed by hybridization at 37°C overnight. For an effective reaction, a buffer is added.
The sample is washed, and unbound nucleic acids are removed and placed in the detection chamber to detect the signals.
A computer-generated software analyses the signals and interprets the results accordingly.
DNA-DNA hybridization:
DNA-DNA hybridization uses the same principle as explained above. Here hybridization occurs between two DNA strands, one is our target and another is a probe which is a 20 to 30 nt long known sequence.
For Example, Southern hybridization, qPCR probes and FISH.
DNA-DNA hybridization is often employed for the purpose to find variations, mutations or any alterations within the sequence. A mutation-specific labeled probe hybridizes and gives signals only if the mutation is present.
DNA- DNA hybridization techniques:
Here in this section, I will elaboratively explain each technique but not in detail as many of them are already there on our blog. I will give a link when required.
Southern blotting:
Southern blotting is a classic and we can say the first-generation hybridization technique. It was originally developed by Southern E in 1975 and from there it got its name. Technically, it is a DNA-DNA hybridization technique.
Digested DNA fragments are allowed to hybridize with known DNA probes on a nylon sheet or membrane. When exposed to an X-ray film the hybridization can be investigated. Such a technique was used to study DNA alterations in the past.
qPCR:
Quantitative PCR uses both hybridization and amplification. In principle, a known sequence probe (flour labeled) binds at its complementary location. The probes of qPCR are specially designed and contain not only a fluorescent label but also a quencher dye too.
The progression of amplification detaches the probe, unquenches it and releases fluorescence. Signals denote a positive amplification reaction. qPCR-based techniques are used in many fields including diagnosis, research and prognosis of the disease.
It can quantitatively measure the amount of DNA present.
Related article: Real-time PCR: Principle, Procedure, Advantages, Limitations and Applications.
FISH:
FISH is fluorescence in situ hybridization. It’s also a DNA-DNA hybridization-based technique that can directly locate any sequence on a chromosome. In principle, labeled- probes are allowed to hybridize with metaphase chromosomes.
If it finds the complementary location, hybridization occurs and is visualized under a microscope. FISH belongs to the molecular cytogenetic technique. It can find larger deletions, duplications and inversions directly on chromosomes.
It is often used for genomic mapping as well.
Related article: A Brief Introduction to (FISH) Fluorescence In Situ Hybridization.
DNA-RNA hybridization:
This also uses the same principle but the target is DNA and the probe used is RNA sequence, complementary to the DNA. An example of DNA-RNA hybridization is Northern blotting and RNA microarray.
RNA microarrays are used to study and determine gene expressions.
DNA-RNA hybridization techniques:
Northern blotting:
Northern blotting works on the same principle of DNA hybridization and southern blotting but here instead of DNA, RNA is used as a template. Both DNA or RNA, labeled probes are used for the hybridization depending upon the requirements.
Hybridization occurs on a nylon sheet or membrane while results are analyzed using X-ray film. Northern blotting is used for RNA studies and gene expression. However, it is now replaced by microarray.
Microarray:
Microarray is a robust, accurate and very precise DNA-RNA hybridization technique that is capable enough to screen thousands or millions of gene expressions or alterations.
Basically, for gene expression studies RNA extraction is performed followed by cDNA preparation. Now the cDNA is allowed to hybridize with known sequence RNA probes immobilized on a glass slide. Probes when finding complementation, hybridize there and give a signal.
Applications of DNA hybridization:
Any hybridization technique can
- Study a sequence
- Finds sequence variations- deletions, duplications, insertions or translocations.
- Study gene expression.
- Used in phylogeny and chromosome mapping.
Limitations of DNA hybridization:
Hybridization has serious shortcomings.
- It is a laborious and time-consuming process.
- It is more error-prone as it can give false hybridization signals by partial or incomplete complementation.
- It can mislead the results.
Wrapping up:
I personally like DNA hybridization. In fact, in my PhD research, I selected a microarray for investigations because it provides ease and at the same time can study millions of alterations. Unfortunately, the flaws are too serious.
And on the other hand, amplification and sequencing-based assets are way more powerful, accurate and precise. So we have to use it. Still, FISH is unmatched and used in diagnostics for the screening of cancer.
There are many ready-to-use FISH probes and kits for various cancer-associated alterations available on the market. Nonetheless, high-end expertise and experience are required for hybridization-based analysis.
Sources:
Fisher S. Not just “a clever way to detect whether DNA really made RNA”: The invention of DNA-RNA hybridization and its outcome. Stud Hist Philos Biol Biomed Sci. 2015 Oct;53:40-52. doi: 10.1016/j.shpsc.2015.07.002. Epub 2015 Jul 25. PMID: 26209888.


