“NGS is a high-throughput sequencing platform that can sequence the entire genome using the massive parallel approach. Learn the basic concept of NGS in this article.”
Sequencing is becoming increasingly popular in research, diagnosis and even in academics. We meet students live every week, in our weekly free live class series. They are more interested in learning sequencing.
And why not!
Sequencing technologies are shaping our future as they are penetrating into diverse fields, rapidly. Scientists want sequence-level information and so do doctors! To understand the molecular mechanism behind a disease or health condition.
Two techniques ruling the sequencing industry are the Sanger sequencing and the NGS. Where the advancements and automations in Sanger sequencing make it a gold-standard method, NGS is rapidly gaining popularity too.
NGS is a fascinating and entirely new ‘word’ for students. Only those who worked with NGS technology can explain it comprehensively at a technical level. I have years of experience working in a genetic lab and have even undergone extensive NGS training.
In this article, I will simplify NGS for not only the students but also for those who want to know this technique, its technical procedure, advantages, limitations and chemistries used by various platforms.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy to understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Key Topics:
What is NGS?
NGS stands for Next-Generation Sequencing. It is not a ‘single instrument’; it is a complete technology that allows high-throughput sequencing and analysis. For instance, Illumina and Ion Torrent are pioneers in NGS technology, however, other companies also have a significant presence.
Among various sequencing generations, previous generation sequencing can effectively sequence a single gene or DNA fragment of approximately 500 to 1000 bp, for instance, Sanger sequencing.
NGS, on the other hand, uses the massive parallel sequencing approach. Instead of a single fragment of a few hundred base pairs, the entire genome can be simultaneously sequenced in a single experiment.
This doesn’t sound impressive at first glance, but let me tell you it took almost 13 years to sequence the complete human genome that an NGS machine can do within 24 to 48 hours.
Now, it sounds fascinating! Isn’t it! That powerful present technology is!
But it doesn’t work as you think. I know what you are thinking; it will sequence just like the Sanger sequencer does! Read the complete genome, base-by-base, right?
Wrong!
The massive parallel approach is entirely different from the conventional sequencing procedure. We first need to fragment the genome, make it accessible for the machine to process it and then sequence each fragment, many times.
That we will cover in the procedure part, in detail.
Historically, the story began with the introduction of pyrosequencing technology in the early 2000s. With a complex and tedious sequencing procedure, pyrosequencing introduced the concept of high-throughput sequencing. And hence, it was referred to as the first NGS technology.
You can read our previous comprehensive article on pyrosequencing to gain more insight into the technical part.
Coming to the point.
Roche acquired pyrosequencing technology and introduced Roche 454 pyrosequencing later on. Companies understood the importance of throughput in the sequencing industry. And afterward, in a very short time, SOLiD sequencing, Illumina and Ion Torrent sequencing platforms came into the market.
All these technologies are collectively known as Next-Generation Sequencing.
So now you know! These are different machines, manufactured by different companies, work on different chemistries and may have different but high throughput.
We discussed the history of DNA sequencing in this article. Click the link to read more.
Definition of NGS:
Next-generation sequencing is a high-throughput sequencing technology that works on the principle of massive parallel sequencing and can sequence the complete genome of an organism.
Principle of NGS:
NGS works on the principle of massive parallel sequencing, in which millions of genomic DNA fragments are sequenced by immobilizing them on a solid surface. DNA is fragmented to construct the library and sequenced simultaneously by generating clusters through different chemistries.
Finally, the sequencing file is processed and analyzed to call the variant and generate the report.
Steps in NGS:
We already discussed sequencing and genome sequencing steps in various articles. In this article, we will discuss the steps in the context of NGS. NGS typically involves these steps:
- Nucleic acid isolation
- DNA fragmentation
- Adaptor ligation
- Library preparation
- Cluster generation
- Sequencing
- Data Analysis
Nucleic acid extraction:
To initiate any experiment in molecular genetics, we need a pure and high-yield nucleic acid, either DNA or RNA. From any biological sample, nucleic acid has been extracted, purified and used for analysis.
Manual methods, spin column kits and automated DNA extraction are a few options for nucleic acid extraction.

Note:
In the case of RNA extraction, the RNA integrity number is checked to evaluate the integrity of RNA before downstream processing.
DNA fragmentation:
As discussed, the complete genome can’t be directly sequenced. To make it sequencing-ready, we need to fragment them into sequencable fragments. NGS platforms (Illumina and Ion Torrent) work on short-read sequencing.
The genomic DNA is fragments into 200 to 600 bp long smaller sized fragments, based on the platform requirement. Mechanical shearing, sonication and enzymatic digestion are a few common options used for DNA fragmentation.
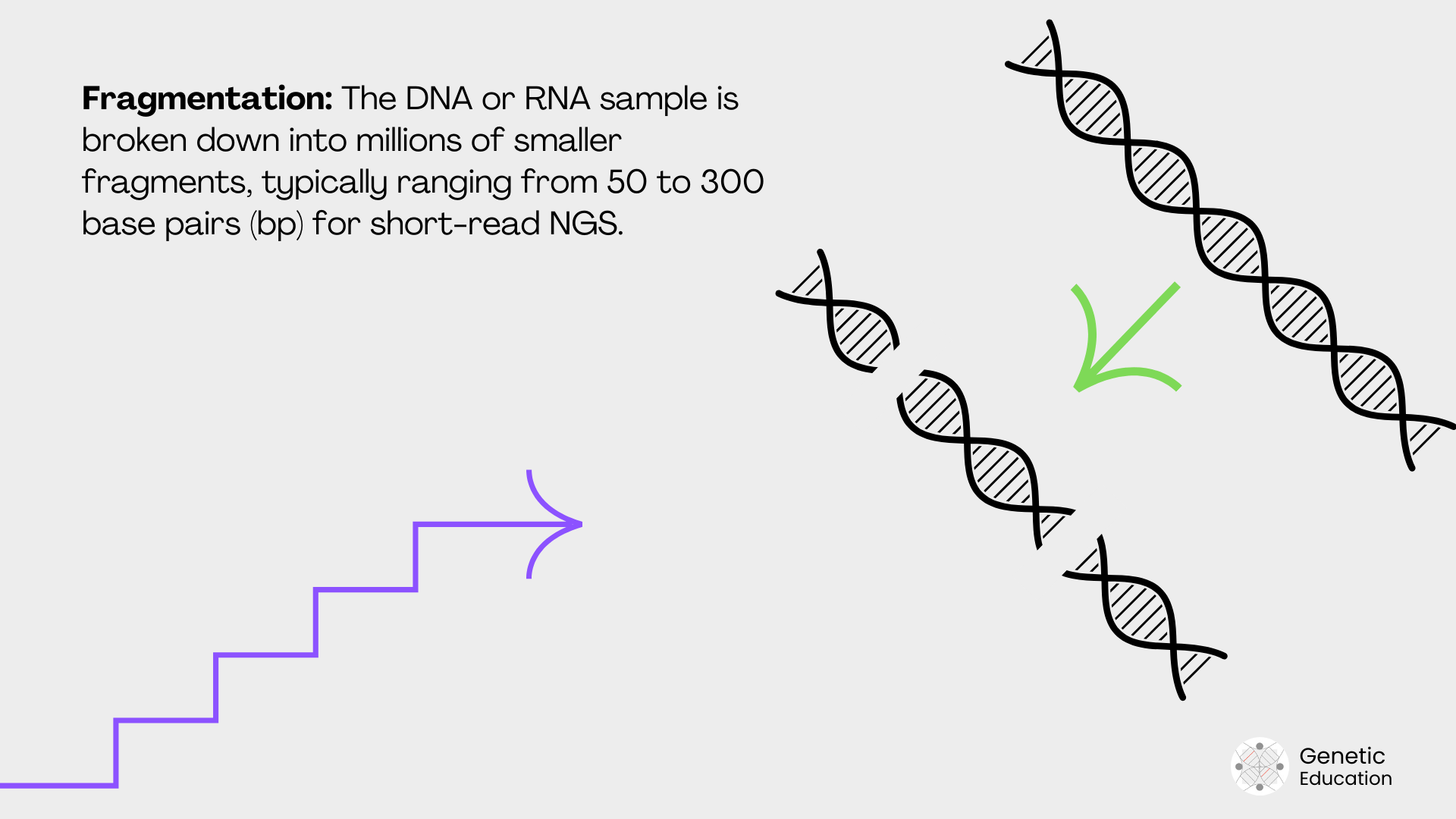
Adaptor ligation:
Now, how does the machine know how the fragments are arranged after the sequencing? To do so, we use ‘known sequence adaptors.’ Adaptors are ligated to each DNA fragment, which attaches the adaptor-specific sequence to the solid surface.
Adaptors help during read assembly. Common kits available for adaptor ligation are T4 ligase-based and transposon-based ligation.
Library preparation:
Libraries are collections of similar-sized fragments. The fragments are size-selected, purified, enriched (if needed) and stored in the library. Library preparation kits and instruments are available.
Illumina uses a high-quality library preparation kit, whereas the Ion Torrent uses the ‘Ion Chef’ instrument for library preparation and enrichment.
Library enrichment is a process in which the fragments are enriched or increased using the PCR amplification to ensure adequate fragment concentration during sequencing.
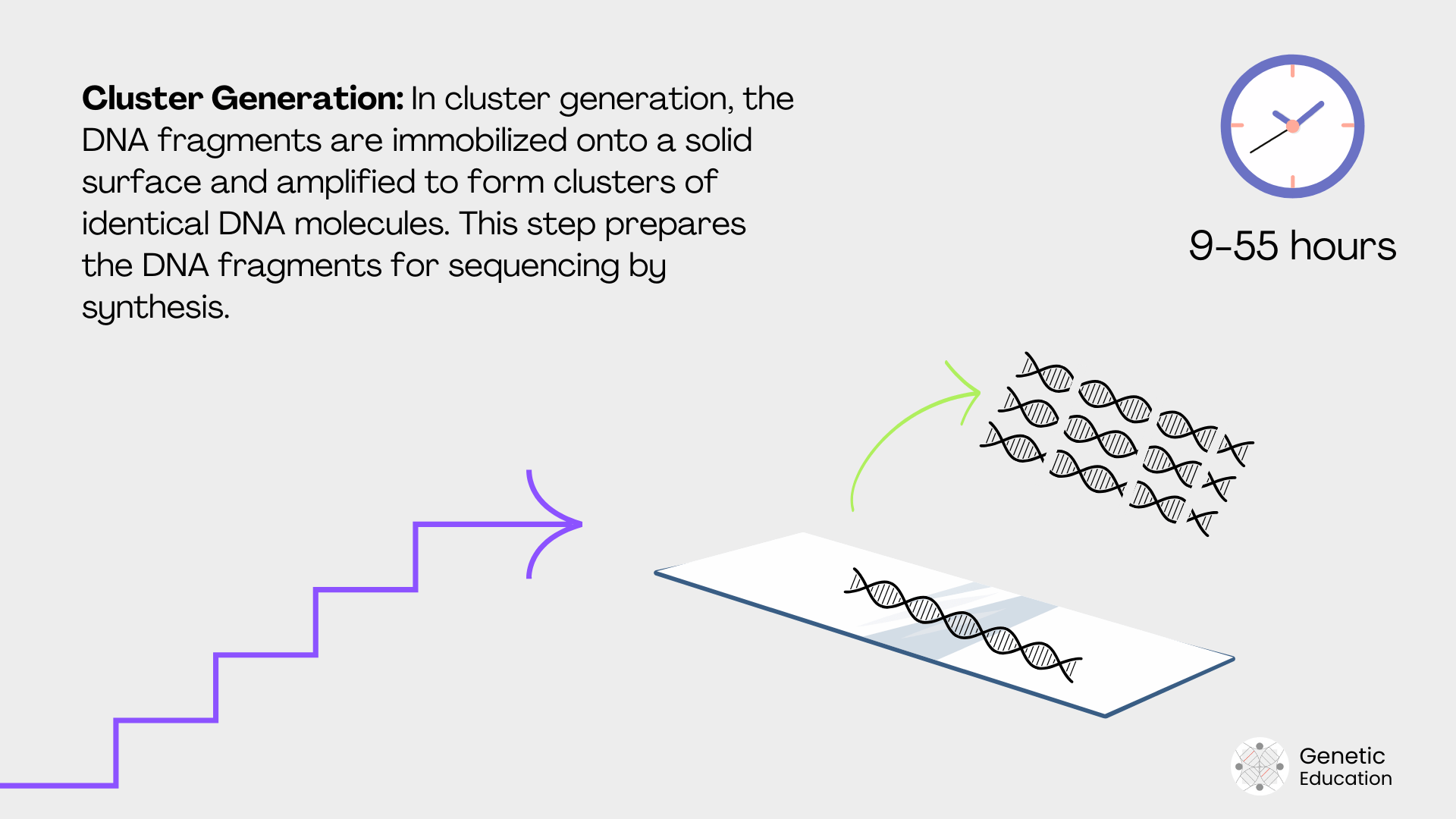
Cluster generation:
Now, when we apply the library to the solid surface, each DNA fragment binds with the adaptor-specific sequence. Afterward, based on the chemistry of the instrument, the cluster of fragments is generated by amplification.
Illumina uses bridge amplification, whereas Ion Torrent uses emulsion PCR for cluster generation.
Sequencing:
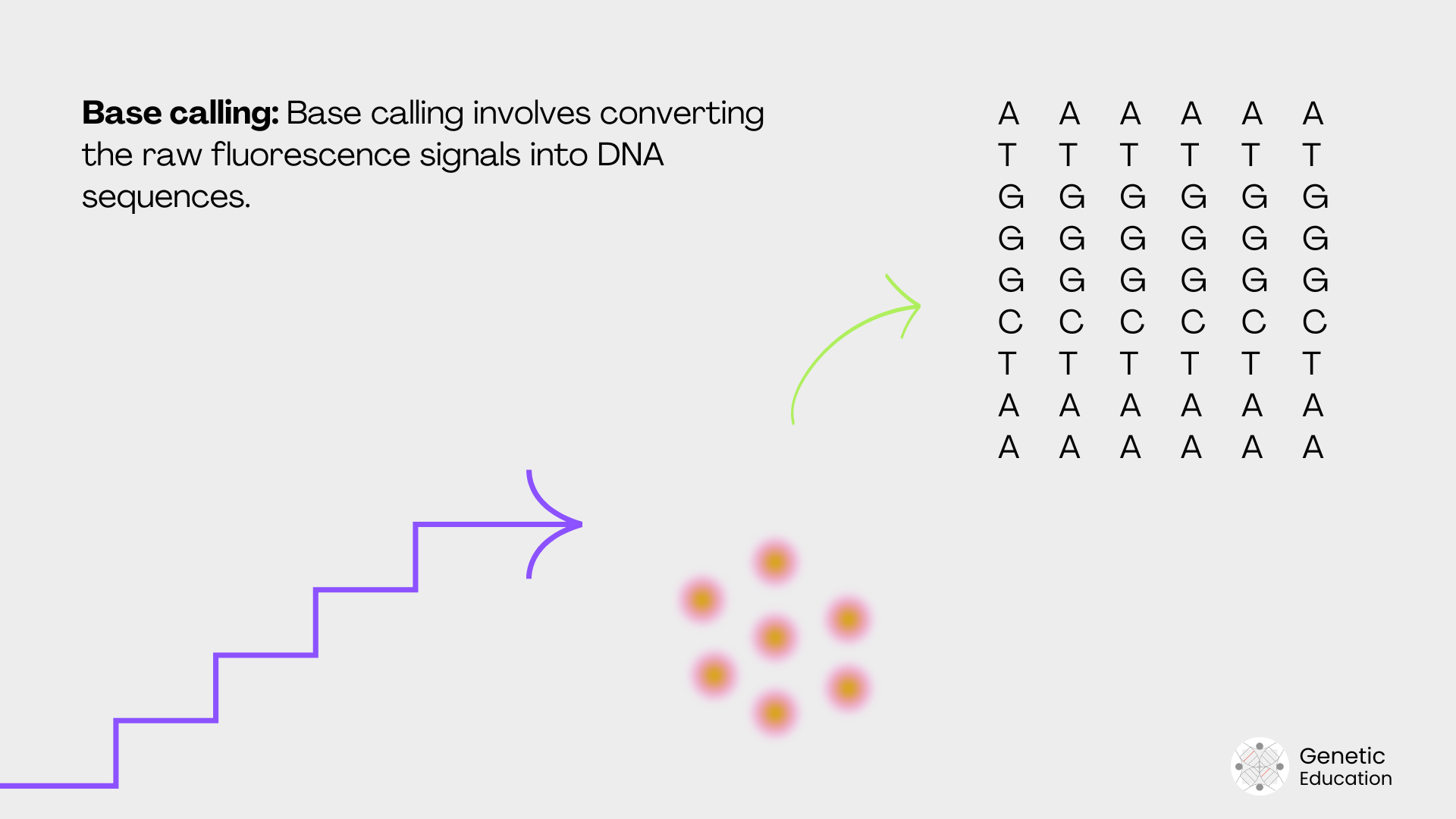
Simultaneously with the cluster generation, the machine also performs sequencing. The detection system captures the signal during amplification or base incorporation. Such signals are either fluorescence signals (in the case of Illumina) or H+ ions or pH change (in the case of Ion Torrent).
As aforementioned, the detection system detects every base, one after another, from each fragment of the cluster and generates the raw sequencing file.
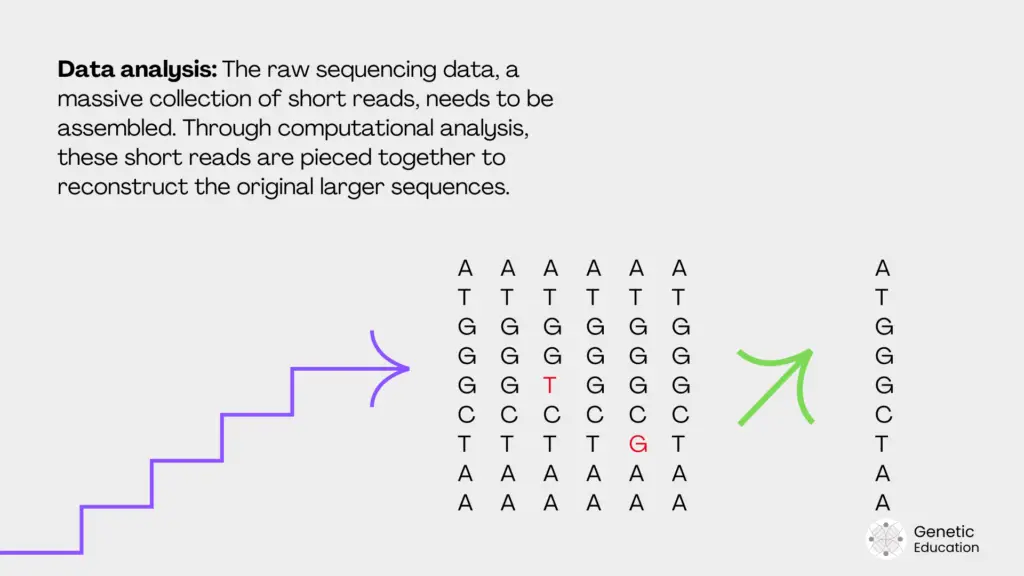
Data analysis:
Now, after completion of the sequencing process, the raw data file is processed for data analysis. Using various bioinformatic tools and pipelines, scientists extract meaningful outcomes from the file.
Meaning, it converts the raw data into interpretable data. This step comprises base calling, genome assembly and variant calling as sub-steps and is explained in the table below.
| Step | Explanation |
| Quality control | Ensures high-quality data by removing errors and artifacts. |
| Read mapping and read alignment | Aligns reads to a reference genome for variant detection. |
| De novo assembly (optional) | Constructs genomes without a reference, useful for new species. |
| Variant calling | Identifies SNPs, indels, and structural variations. |
| Annotation | Adds functional information to identified variants. |
| Pathway and functional analysis | Helps understand the biological significance of genetic changes. |
| Data visualization | Provides a graphical representation for easier interpretation. |
Summary of the NGS steps:
| Step | Purpose | Importance | Options | Time |
| Nucleic acid isolation | Isolate high-quality DNA/RNA. | Ensures purity and integrity. | Manual, spin column, automated extraction. | 1–3 hours |
| DNA fragmentation | Break DNA into small fragments. | Creates optimal fragment sizes. | Sonication, enzymatic digestion, tagmentation. | 10–30 min |
| Adaptor ligation | Attach adapters for amplification and sequencing. | Enables sequencing compatibility. | T4 DNA ligase, transposase-based ligation. | 30–60 min |
| Library preparation | Generate a sequencing-ready DNA library. | Ensures uniform fragment distribution. | Library preparation kits and instruments | 2–4 hours |
| Cluster generation | Amplify DNA fragments for sequencing. | Increases signal detection. | Bridge PCR (Illumina), emulsion PCR (Ion). | 2–6 hours |
| Sequencing | Determine nucleotide sequences. | Produces raw sequencing reads. | Illumina SBS, Ion Torrent, Nanopore. | 6–48 hours |
| Data analysis | Convert reads into interpretable data. | Identifies variants and biological insights. | FastQC, BWA, GATK, bioinformatics pipelines. | Days to weeks |
NGS chemistry:
As aforementioned, the two main players in the NGS industry are Illumina and Ion Torrent. Sequencing by synthesis and semiconductor sequencing chemistries are followed by Illumina and Ion Torrent, respectively.
Sequencing-by-synthesis:
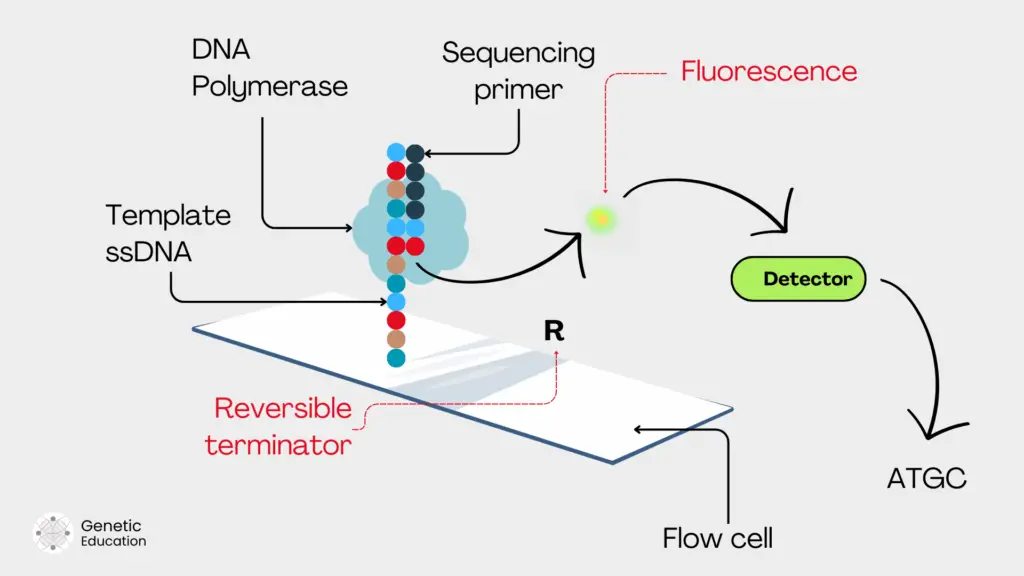
Sequencing by synthesis is the most common and widely used NGS chemistry. It is primarily used in the Illumina sequencing platforms. Once the DNA fragment is immobilized on the solid surface, the SBS begins.
The primer binds with the fragment and polymerase starts incorporating the nucleotide on the fragment. During each round of amplification, the polymerase incorporates a fluorescently labeled complementary nucleotide.
It uses a reversible terminator dye; hence, after fluorescence reading, the nucleotide is released to prevent false signals. The laser emits fluorescence, the camera captures it and the optical detector detects and converts the light signal into a digital signal.
To comprehend your understanding, you can read our in-depth article on this topic. The link is already provided.
Semiconductor sequencing:
Ion Torrent NGS platforms rely on semiconductor sequencing chemistry. It is also a sequencing by synthesis chemistry, but instead of the fluorescence signal, it captures the pH change signals.
Now, here, the fragments are attached to the microtiter wells on a solid surface and follow the bridge amplification. Once the polymerase incorporates the nucleotide, it releases the H+ ion. This hydrogen release induces a pH change, which is detected by the highly sensitive semiconductor and converted into a digital signal.
On the technical side, so many things are involved in the present chemistry that we will cover in a separate article.
NGS Machine and Instrumental setup:
The NGS machine, chemistry and detection system may vary among companies, however, some parts or systems will remain the same in all the machines. In this segment, we will understand the instrumental setup of the NGS machine.
Solid surface, reagent cartridge, fluidic system, temperature control system, detection system, waste management and controller system are common components of an NGS machine.
Let’s understand the role of each, one by one.
Solid surface:
Solid surface, or in simple words, we can say a ‘highly sophisticated slide’ is a surface on which the sequencing clusters are generated. Meaning, the sequencing reaction happens here. This solid surface contains microscopic channels through which the reagents pass and helps the immobilised DNA to amplify.
Flow cell, semiconductor sequencing chip and ZMW (Zero Mode Wavelength) in Illumina, Ion Torrent and ONT, respectively, are the solid surface or chip used for cluster generation.
Reagent cartridge:
The reagent cartridge contains all the necessary reagents needed during the sequencing reaction, such as dNTPs, buffer, DNA polymerase, etc. The reagent cartridge is loaded on the sequencer machine and supplies reagents.
Fluidic system:
The fluidic system is a crucial component of the NGS machine as it controls the flow of reagents and washing fluid during the sequencing reaction. It contains pumps and valves for its function and is controlled by a central software.
Temperature controller:
TCS is required for two common purposes: to control the temperature of the sequencing reaction by heating and cooling steps and to perform denaturation, annealing and extension during the amplification.
Reagents may be inactivated during heat generation, so to maintain adequate temperature during the reaction, the TCS is installed in the machine.
Detection system:
The detection system component varies from chemistry to chemistry. For instance, the Illumina sequencing-by-synthesis chemistry uses a high-definition laser, detector and a high-definition camera to emit the fluorescence according to the base, to capture the fluorescence using optical sensors and to capture the signals, respectively.
In the case of the Ion Torrent sequencer, the detection system consists of a sensitive semiconductor detector that detects a minor pH change during the incorporation of the nucleotides. It detects the pH change in real time.
So, in conclusion, the detection system contains the component to detect the nucleotide incorporation or read the nucleotides from the sequence or during the sequencing process.
Waste management system:
Waste fluids and consumables are generated during the sequencing process. The machine is equipped with an inbuilt waste management system that collects the solution and reagent waste after each round of sequencing.
This helps improve the sequencing results by minimizing the background noise.
Controller software:
Lastly, the controller software is installed in the machine that controls all the operations in the machine. It controls the sequencing progress and monitors the overall reaction, including the reagents needed and consumed, temperature required and given, etc., in the reaction.
Advantages of NGS:
In this segment, we are going to discuss how the NGS is superior or important than other sequencing platforms.
High throughput:
High throughput is the main USP of NGS technology, it can generate millions to billions of reads in a single experiment. Meaning, it is powerful enough to sequence the entire genome of an organism.
Rapid sequencing:
It allows faster sequencing; the massive parallel approach and simultaneous sequencing of fragments can provide sequencing of the entire genome within 24 to 48 hours.
Multiplexing capabilities:
NGS yields a huge amount of data. So by multiplexing various samples in a single experiment (using the index sequences), time and cost can be reduced. It also reduces the TAT.
Automation:
NGS platforms available in recent times are highly automated. Starting from sample preparation of data analysis, all the steps can be performed with minimal human interference. This reduces the error rate and increases the accuracy of the result.
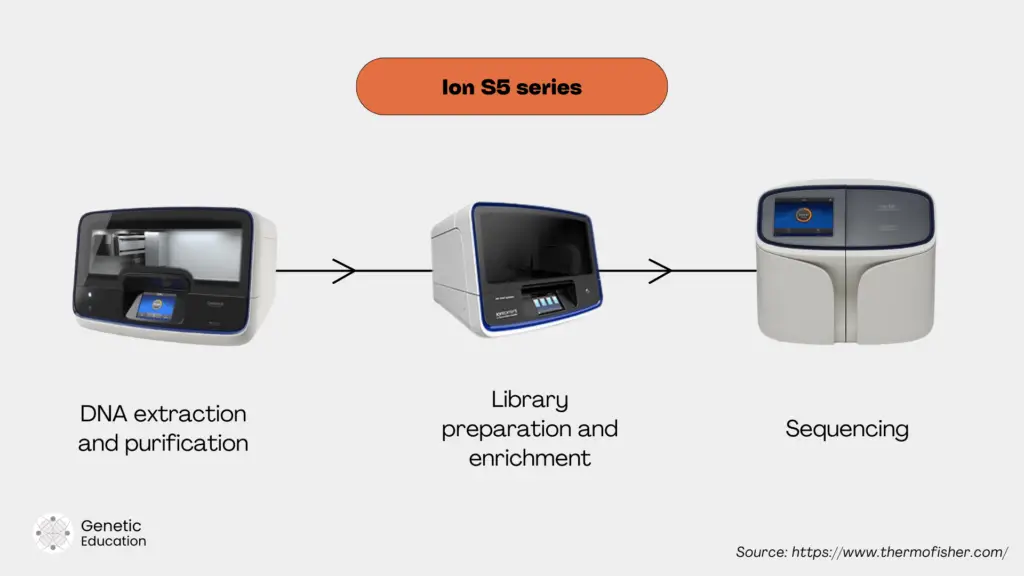
For instance, Ion Torrent’s automated DNA extractor, Ion Chef library preparation and Genexus sequencer are a complete two-touch automated NGS ecosystem that covers DNA extraction, library preparation and enrichment, and sequencing and analysis under a single ecosystem.
Highly accurate:
The integration of short-read sequencing and higher sequencing depth makes the entire NGS platform highly accurate.
Sequencing depth: Also known as sequencing coverage, refers to how many times a particular nucleotide from a fragment is read
Scalable:
NGS testing is in high demand! Various test kits and panels are now available. For instance, a dedicated panel for cancer genes, reproductive medicine, and newborn screening etc. Such advancements make the technology scalable and readily available for testing.
Detect rare variant:
On the technical side, NGS can detect low-frequency or rare genomic variants with high coverage and accuracy, even from a mixed sample. This makes it the most important and versatile technique to use in the diagnostic industry.
In addition, it is suitable for medium to large-sized projects and can also handle low-volume samples, without compromising the accuracy.
Limitations of NGS:
Despite the versatility, high accuracy and throughput, NGS also has several serious limitations. Here are some.
Data-intensive:
NGS generates a huge amount of data that is difficult to store, manage, process and analyze. It required sophisticated servers or cloud storage, computational power and a skilled bioinformatician team to handle the data.
Prone to errors:
It may also produce errors during sequencing. Particularly, in high GC-rich, homopolymeric and repetitive regions. Errors are also reported during data analysis, often.
Costly:
The technology itself is costly due to high initial investment, running costs, and the requirement of a separate bioinformatics ecosystem and infrastructure.
Time-consuming:
NGS analysis has a very high TAT as data processing and analysis are time-consuming and tedious tasks.
Short read limitations:
Although short-read sequencing is extensively used in NGS, it has one major limitation. It can’t resolve larger structural variants. For instance, a homopolymeric sequence block exceeding the read length.
Biases and artifacts:
The present technology highly relies on library preparation and PCR amplification which can introduce biases and artifacts during the sequencing. Such problems make the overall analysis procedure difficult and hinder the assembly process.
Expertise dependency:
NGS requires trained and experienced bioinformaticians and IT professionals for handling the dry lab work analysis.
Wrapping up:
NGS is a revolutionary sequencing platform, used in nearly all the biological science fields in recent times. Its power to sequence a complete genome allows researchers to gain meaningful insight from their experiment.
This provides new knowledge and generates novel genomic information. However, the higher cost, computational and bioinformatics requirements restrict its full potential. Nonetheless, the output that it provides is more lucrative.
NGS also changes the clinical field, in particular, oncology. Due to its wide range of applications, we will write another article exploring NGS’s applications in various fields.
I hope this beginner-friendly article will strengthen your sequencing knowledge and clear your NGS basics. In addition if you want to learn more, you can enroll in our basics to advanced NGS course.
Sources:
Rodriguez R, Krishnan Y. The chemistry of next-generation sequencing. Nat Biotechnol. 2023;41(12):1709-1715. doi:10.1038/s41587-023-01986-3.
Rodriguez, R., Krishnan, Y. The chemistry of next-generation sequencing. Nat Biotechnol 41, 1709–1715 (2023). https://doi.org/10.1038/s41587-023-01986-3.
Behjati S, Tarpey PS. What is next generation sequencing?. Arch Dis Child Educ Pract Ed. 2013;98(6):236-238. doi:10.1136/archdischild-2013-304340.