Between the microarray vs RNA seq, RNA sequencing allows scientists to investigate novel RNA variants whereas microarray can investigate known sequence variants effectively. Henceforth, it is important to understand which technique to use when.
Let’s explore some of the technical differences between microarray vs RNA seq.
“Transcriptomics” is a study of understanding the total mRNA of a cell or the expression of all genes. It is an expressed pool of genes, demonstrating how many genes are expressed, and how many times! It is vital for understanding the proteomics part.
RT-PCR, RNA sequencing and microarray-based techniques are now available to study gene expression. Such techniques are state-of-art, high throughput and robust. However, RT-PCR can quantify a single or a few mRNAs whereas sequencing and microarray can study thousands of mRNA or the entire transcriptomics.
So it is crucial to select a technique as per the requirement of the assay, indeed. This article contains information that helps you to make a decision, therefore this exploration is more commercial, opinion-driven and has decision-making intent rather than informational.
I will explain both the techniques and some common similarities and differences between microarray vs RNA sequencing.
Key Topics:
What is an RNA microarray?
Microarray is a hybridization and chip-based, quantitative, transcriptomic analysis technique. On a technical side, the assay contains a microscopic chip made up of transcriptome-specific probes.
Probes are short, 20 to 30nt long known oligonucleotides, immobilized on a glass slide to prepare a chip. RNA sample is selectively applied on the chip and allowed to hybridize with probes.
The presence of fluorescence molecules helps to detect and study the results. Note that case and control samples are applied to hybridize using two different dyes or fluorochromes. Microarray is an easy, robust, reproducible and sensitive transcriptomic analysis technique.
What is RNA sequencing?
It is a sequencing-based assay that simply reads the sequence of a transcriptome. The total mRNA is reverse transcribed into cDNA and arranged into cDNA libraries. If you want to learn more about cDNA libraries, read this article: The Process of cDNA Synthesis and cDNA Library Preparation.
Now each fragment from a library is read by the machine, detected, data collected and interpreted using computational software.
RNA sequencing is also a state-of-art, high throughput, robust and reproducible transcriptomics analysis technique.
Both techniques can somehow do the same work, so what are the differences? Let’s find out.
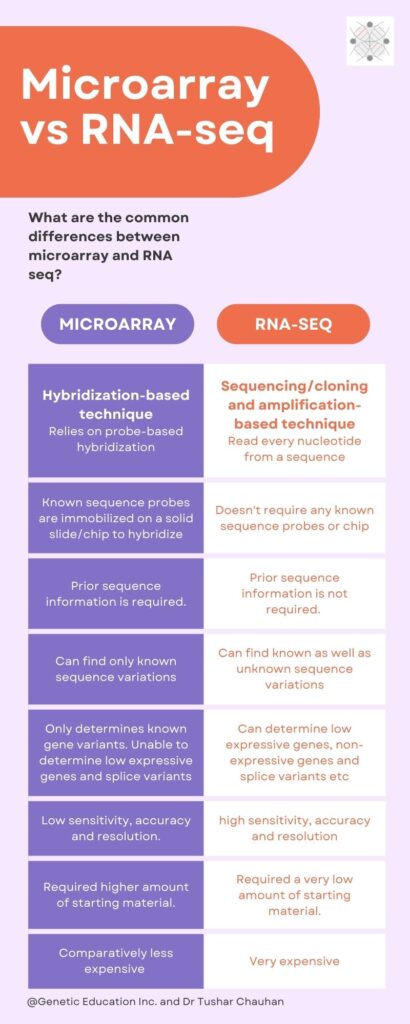
Microarray vs RNA-seq- Differences:
Microarray is a hybridization-based technique while RNA-sequencing is a sequencing-based technique.
Microarray uses a microscopic chip on which known transcriptomic sequence-specific probes are immobilized whereas RNA-seq doesn’t use any chip or transcriptomic-specific probes.
So technically, microarray requires known sequence probes for analysis while RNA-seq doesn’t need such types of probes.
The principle of microarray relies on the hybridization of probes with the sequence by following strict hybridization conditions, while the principle of RNA-seq relies on the amplification or enrichment of the sequencing followed by detection using fluorescent tagging.
Steps in microarray are RNA extraction, reverse transcription, fragmentation, hybridization, washing and detection, while the steps in RNA-seq are RNA extraction, reverse transcription, fragmentation, cDNA library preparation, library enrichment, sequencing and detection.
Importantly, microarray requires previous sequence information for preparing probes whereas RNA-seq doesn’t require any sequence information. Henceforth, here is the catch, sequence information is a prerequisite for microarray but not for RNA-seq.
So it is very crucial to know that the microarray analysis only allows us to investigate or study known sequence/transcriptomic variants. On the other side, we can find new sequence variations as well as study the existing ones using the RNA-seq technique.
I also want to add an important technical point from my side. The microarray can only determine copy number variations whilst RNA-seq can find any CNV as well as SNPs too. Microarray can’t find any known or unknown SNPs.
From an application point of view, unlike the microarray, RNA-seq can study SNPs, indels, CNVs, gene fusion and sequence rearrangements.
Whereas
From a detection point of view, microarray comes up with a straightforward and easy detection tool, while to study RNA-seq variations, one needs extensive bioinformatics setup, experienced personnel and trained bioinformaticians.
Microarray has limited resolution and detection range, it can’t detect low expressive genes effectively, contrary, sequencing can effectively detect low expression genes too.
So in comparison to the microarray, RNA-seq has high sensitivity, specificity, wide-detection range and high accuracy.
It also denotes that, dissimilar to the microarray, RNA-seq can find and quantify low abundant transcripts which eventually helps determine accurate transcriptomics analysis.
As per my personal experience, RNA-seq in fact, any NGS-based sequencing technology has low or minimal background signal-to-noise ratio. This means, it’s more powerful and accurate, in disaccordance, microarray can give false hybridization, cross or no-hybridization.
Close comparison:
| Microarray | RNA-seq |
| Low dynamic range | Broad dynamic range |
| Can’t do transcriptomes without any reference genome. | Can do transcriptomes without any reference genome. |
| Can’t detect unannotated genes | Also detect unannotated genes |
| Can’t detect novel alterations | Detects novel alterations |
| Can’t detect alternative splicing events and alterations at a sequence level. | Detect alternative splicing events at a sequence level. |
Despite the fact that the RNA-seq technology has great value in transcriptomics, it has several serious shortcomings too. It generates a huge amount of data which is indeed complex, tedious and hard to manage and process.
It requires extensive bioinformatics parts whereas microarray generates comparatively less data and is manageable by built-in software.
RNA-seq is newer and emerging and thus has limited standardized protocols and optimization scopes. On the other side, the hybridization-based microarrays are well penetrated and contain a huge amount of research and scientific data.
In terms of cost, both techniques are equally very costlier, but the microarray is a bit cheaper than the RNA-seq. See sequencing works on a totally different platform and needs extensive dry lab analysis so companies further have to pay for that.
And the extra cost of RNA-seq is justified too because ultimately it would give us more, novel, unexplored data regarding the transcriptome.
A typical single microarray costs around $100 to $350 whereas a typical RNA-seq costs around $1000 to $6000 per run which even depends on the technique, chemistry and add-on ordered. For example, simple or NGS, single or paired-end sequencing, long or short-read sequencing etc.
Also, keep in note that this price is for testing, laboratories charge extra for computational, statistical and bioinformatics.
Way back in 2016, I ordered a microarray for 20 samples which cost me 22,000 INR ($278) per sample. Now you have a common question, Should I have to order a microarray or RNA-seq? This common question may help you to make a decision.
- If you want to study a pool of known transcriptomic variants?
- If you want to study known copy number variations?
- If you do not want to want to complicate your experiment and at the same time manage things on a tight budget,
Go with a microarray.
- If you want huge data?
- Want to find unknown variants, SNPs and CVNs of unknown location?
- Want to study splice variants and unexpressed transcripts?
- Want high resolution, sensitivity and accuracy?
You can go for RNA-seq.
I highly recommend you to take the advice of your professor, expert or guide, discuss your vision or results you want and then go further.
Similarities:
Both techniques are majorly different, despite the fact that there are several similarities between them as well.
Both are used to study transcriptomics, thereby gene expression.
Starting material for each assay must be RNA and need the reverse transcription to convert it into cDNA.
The instruments for both are expensive and require a separate setup.
Both techniques are highly reproducible, robust and sensitive.
Wrapping up:
Sequencing is, nonetheless, by fast the best method till date. But at the same time, it is costlier too and oftentimes, it is not needed. Although it can identify novel alterations, there are still so many known alterations that are still not studied on a huge number of samples.
You can use microarray to study known variations on a large number of samples and find common and uncommon mutations or pathogenic variants related to a gene and you can even do many other things with known sequence microarray.
After all, a chip of microarray can investigate millions of alterations in a single reaction. So by taking such parameters into consideration, design your experiment or order the technique. I hope this article will help you.