“The PCR reaction buffer is enriched with the PCR enhancers that increase the overall reaction performance, some of them are Tris, EDTA, MgCl2, KCl, Formamide, DMSO, TritonX100, Nonidet P40, twin20, 7-deaza-2′-deoxyguanosine 5′-triphosphate and bovine serum albumin.”
“The PCR reaction buffer is the composition of different PCR enhancers that increase the efficiency and the specificity of the PCR reaction”.
During our PCR reaction, we are facing problems of non-specific bindings, reaction failure, primer-dimer, unwanted amplification, and low amplification. To overcome these problems PCR reaction buffer is used.
Some of the major reasons for reaction failure are long primers, high GC content in template DNA, unpurified template, PCR conditions, and concentration of the chemicals used in it. However, such conditions are overcome by using a specific PCR buffer or required component.
But you may wonder what are those chemicals often used as PCR enhancers? Let us find out. In this article, the major focus will be on the different components used in the PCR reaction buffer.
Key Topics:
MgCl2:
If you want to master the PCR technique, you should learn how to use MgCl2 wisely in different reactions. The MgCl2 is a major PCR enhancer present in the Taq DNA polymerase buffer. The enzyme Taq requires the Mg2+ cofactor to start the catalytic reaction and work efficiently. 0.5mM to 5.0mM MgCl2 can be utilized in PCR reaction. However, the concentration of the MgCl2 is depended on the condition of PCR.
Further, the Mg2+ of the MgCl2 binds to the PO3- (phosphate backbone of DNA) and increases the melting temperature of the reaction. This will facilitate more time for the primer to bind at its specific location.
A higher amount of amplification can also be achieved using MgCl2. Nonetheless, too much MgCl2 decreases the specificity of the reaction and increases the non-specific bindings possibilities. How? For that read the article: The Role of MgCl2 in PCR Reaction. This in-depth article will clarify all your fundamentals on how to use MgCl2 in PCR.
On the downside, a higher concentration of MgCl2 decreases the rate of denaturation of template DNA by increasing the melting temperature.
KCl:
KCl facilitates the elongation of the Primer-DNA complex. The K+ ion of the KCl binds to the DNA backbone (same as Mg2+). This binding decreases the electrostatic repulsion between two DNA strands and facilitates the elongation preferentially.
An increased concentration of KCl also increases the power of denaturation, especially for the shorter DNAs (up to 1000bp). Its performance is disappointing for the longer DNA. In long-range PCR lower concentration of KCl is recommended. The final reaction concentration of the KCl in the PCR reaction is between 25mM to 100mM.
Tips
For PCR of 2kb to 10kb DNA fragment use KCl >25mM (you can also avoid it)
Related article: 50 Powerful Applications Of PCR.
DMSO:
DMSO is a major game-changer in the PCR reaction for the template with high GC content. To amplify the DNA with high GC content is actually difficult. The chance of secondary structure or hairpin formation is higher. The DMSO has the power of disrupting the hydrogen bond between the bases which increases the denaturation rate of high GC rich template.
DMSO prevents the hairpin formation by interrupting between two DNA strands and prevents reannealing. Also, it increases the binding specificity of the primer to the template DNA.
Majorly, DMSO can be preferred into the PCR with GC content more than 55%. Use 4% to 10% DMSO into the PCR reaction buffer. Commonly, 5% of DMSO concentration is advisable for a typical complex DNA template.
Read an in-depth explanation of the role of DMSO in PCR reaction here: Role of DMSO in PCR reaction.
Glycerol:
Glycerol is another reagent in the family of chemicals which reduce the secondary structure formation. The glycerol is very impactful and works efficiently because it does not have any side effects in the PCR reaction. 6% of glycerol is recommended.
Formamide:
The formamide is less known for its role in PCR enhancement. It is the best alternative for DMSO. Formamide increases the PCR amplification efficiency of the GC rich region too. A concentration of 1% to 10% is preferable.
Notably, as the formamide can easily form primer-dimer, use it if needed and based on the GC content of the template.
7-deaza-2′-deoxyguanosine 5′-triphosphate:
Once the DNA template denatured, no hydrogen bonds are present between bases. In that case, especially, in a long DNA template, the DNA binds with each other and forms the secondary structure or hairpin structure.
The secondary structure or the hairpin stabilized the ssDNA and hinders in elongation of the DNA. 7-deaza-2′-deoxyguanosine 5′-triphosphate is another chemical that prevents the secondary structure formation into the denatured single-stranded DNA.
It prevents false results in the PCR. The approximate concentration of dc7 GTP is ~40µM.
Betaine:
Betaine, N,N,N-trimethylglycine is a minor PCR additive that prevents the secondary structure formation especially, into the high GC rich region. It surely increases the efficiency and yield of PCR.
Nonetheless, the exact role of betaine in amplification is controversial and not understood properly. Here in combination with the DMSO, betaine increases the specificity of long-range PCR reactions. The ideal concentration of betaine in PCR should be between 0.5M to 3.0M.
Bovine serum albumin:
DNA extracted from the samples such as marine water, soil, freshwater or feces samples contains a high level of PCR inhibitors such as fulvic acid, humic acid, tannic acid FeCl3, and others.
These inhibitors can hinder in the elongation of the newly grown DNA strand. BSA, a natural protein can prevent the PCR reaction from such types of inhibitors.
The viscous nature of BSA prevents the sticking of PCR reagents on to the wall of the PCR tube thereby increasing the surface for reaction to occur. The concentration of bovine serum albumin should be between 300 ng/μl to 600 ng/μl into the PCR reaction.
T4 gene 32 protein:
This protein also prevents the PCR reactions from the inhibitors, however, which types of inhibitor it blocks still not known well. The ideal concentration of T4 gene 32 protein should be ~100 ng/μl.
Non- ionic detergents:
Detergents are normally used in DNA extraction. Common detergents such as Triton X100, Nnonidet P40, Tween20 and SDS are majorly involved in the cell lysis.
However, apart from SDS and CTAB, other non-ionic detergents like Triton X100, Nonidet P40, and Tween20 are great PCR enhancers. Using these detergents in the PCR reaction buffer can greatly increase the yield and efficiency of the PCR reaction.
The detergents can suppress the activity of other contaminants such as SDS, phenol and other DNA extraction buffer traces. It neutralizes the inhibitory effects of contaminants.
However, the excess concentration of non-ionic detergents can inhibit the PCR reaction. 1% to 2% of non-ionic detergents are sufficient and can be used in the PCR reaction buffer
Tetramethylammonium chloride:
It can also increase the efficiency of the GC rich PCR reaction. However, it is less known for its role in the PCR reaction buffer, still, you can use it 100mM in reaction buffer.
Additionally, it increases the specificity of the primer and DNA hybridization and does not allow any non-specific bindings.
Polyethylene glycol:
Polyethylene glycol is less known for its role in the PCR reaction buffer. PEG is the excellent enhancer of the PCR reaction. It works differently than other chemicals. It prevents the re-annealing of denatured single-stranded DNA by binding with the ssDNA.
However, it does not interrupt in Taq polymerase activity, it works like a single-stranded binding protein (prevent annealing of DNA during replication).
Interestingly, it blocks the unbounded primers and prevents dimer formation during the PCR reaction. For efficient PCR reaction buffer preparation use PEG between the concentration of 5% to 15%.
Gelatine:
It is viscous in nature which prevents sticking of Taq DNA polymerase and other PCR reaction reagents to the wall of the PCR tube. Doing this provides more surface area for each chemical to work efficiently.
Also, it prevents excess evaporation during extreme heating in the PCR. The function of gelatine is somewhat similar to the BSA. The recommended concentration is 0.01% to 0.1%.
Bonus Tips: while working on the samples of ancients tissues or ancient DNA and DNA samples having more melanin, use Bovine serum albumin. The concentration of BSA is given above, this will increase the possibilities of PCR results.
The above-enlisted chemicals are PCR enhancers which can be used into the PCR reaction buffer, yet, some other chemicals which are as important as the PCR enhancers. These chemicals are used to maintain the pH and the stability of the reaction.
Tris and EDTA are two important ingredients that serve the purpose of stabilizing the reaction. The DNA is pH sensitive, a constant pH environment between 7.8 to 8.5 is required for DNA to work efficiently which stabilizes the reaction.
Tris-HCl:
Tris-HCl with pH 8.0 maintains the stability of the reaction and constant pH throughout the PCR reaction. This prevents the degradation of the DNA molecule.
EDTA:
EDTA is a chelating agent that blocks mg2+ binding with the Taq DNA polymerase and reduces the polymerization activity. If Mg2+ is not available then how Taq DNA polymerase can work?
The question is valid because if we use EDTA, it definitely suppresses the activity of Mg2+, but it can also enhance the reaction.
Using EDTA into PCR will chelate the excess unused Mg2+ ions that might inhibit the PCR, also, it chelates other ions presents into the PCR reaction mixture. Therefore, if we use EDTA in a lesser proportion of MgCl2, it will enhance the PCR reaction.
Tris and EDTA are two major components that are required in each and every step of performing any experiments with DNA.
Summary of the PCR reaction buffer:
GC rich template DNA: use 7-deaza-2′-deoxyguanosine 5’-triphosphate, betaine, DMSO, formamide, glycerol, and Tetramethylammonium chloride.
Prevent secondary structure formation: formamide, DMSO, glycerol, betaine and 7-deaza-2′-deoxyguanosine 5’-triphosphate
Increase primer binding efficiency: MgCl2, KCl and Tetramethylammonium chloride
Increase Tm: Tetramethylammonium chloride
Decrease Tm: DMSO
Enhance Taq polymerase activity: MgCl2, polyethylene glycol and gelatine.
My ultimate guide for optimizing PCR reaction buffer:
The condition of any PCR reaction varies from lab to lab and person to person. The protocol that I design, might not work properly for you and it is very often in molecular biology.
Though we have given concentration for all the different components which are used in the PCR reaction buffer preparation, you have to optimize or standardize your own protocol into the lab.
How?
Let me explain. Let’s take the example of MgCl2. Our aim is to optimize the MgCl2 in our reaction.
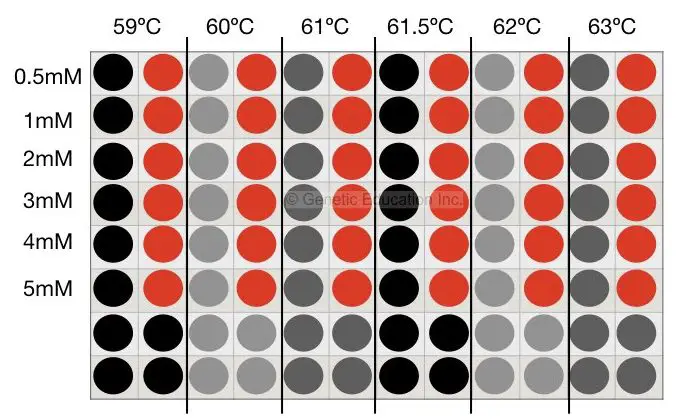
Six different temperature gradients are there in our gradient-conventional PCR machine each with the minimum temperature difference of 0.5°C. First, we will find out the annealing temperature for our PCR reaction,
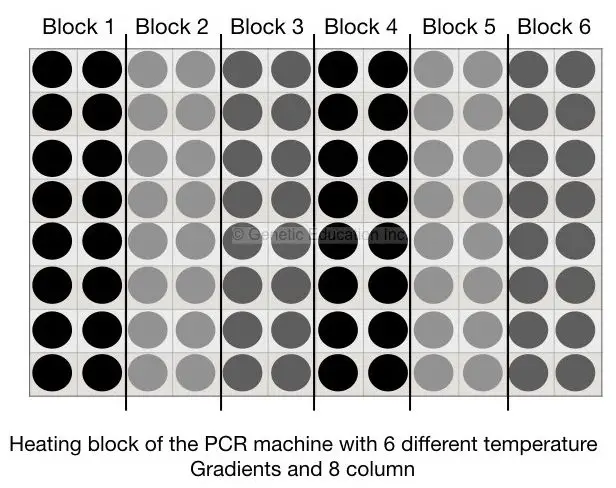
Mix all reagents as per the protocol, suppose our annealing temperature (by primer 3 software) is 61°C. Now design 6 different tubes and put is in the PCR machine. Set the temperature ranging from 59°C to 63°C (59°C, 60°C, 61°C, 61.5°C, 62°C, 63°C). The condition is shown int to the figure below,
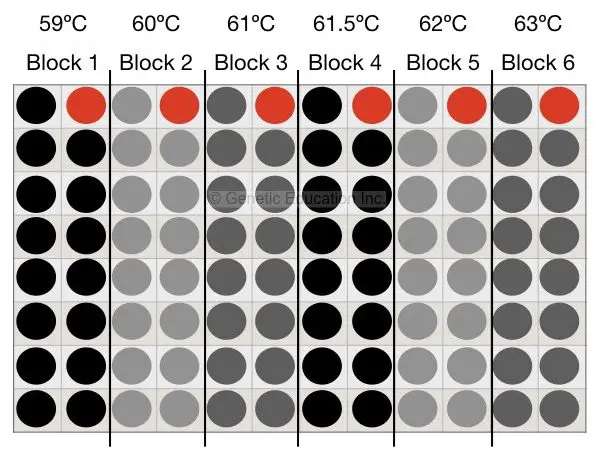
After agarose gel electrophoresis, suppose we get some amplification at the temperature 62°C. Now we have to enhance that PCR reaction results.
Prepare another set of reactions as per the protocol. Add MgCl2 0.5mM to 5 mM.
Tube 1- 0.5mM
Tube 2- 1.0mM
Tube 3- 2.0mM
Tube 4- 3.0mM
Tube 5- 4.0mM
Tube 6- 5.0mM
Place it into the PCR machine at 62°C temperature analyze the results on 2% agarose gel. Or you can follow another method which is tedious but quite accurate, the method is shown in the figure below,
Finally, we are giving you our recipe of the PCR reaction buffer, what components we are using in preparing the PCR buffer.
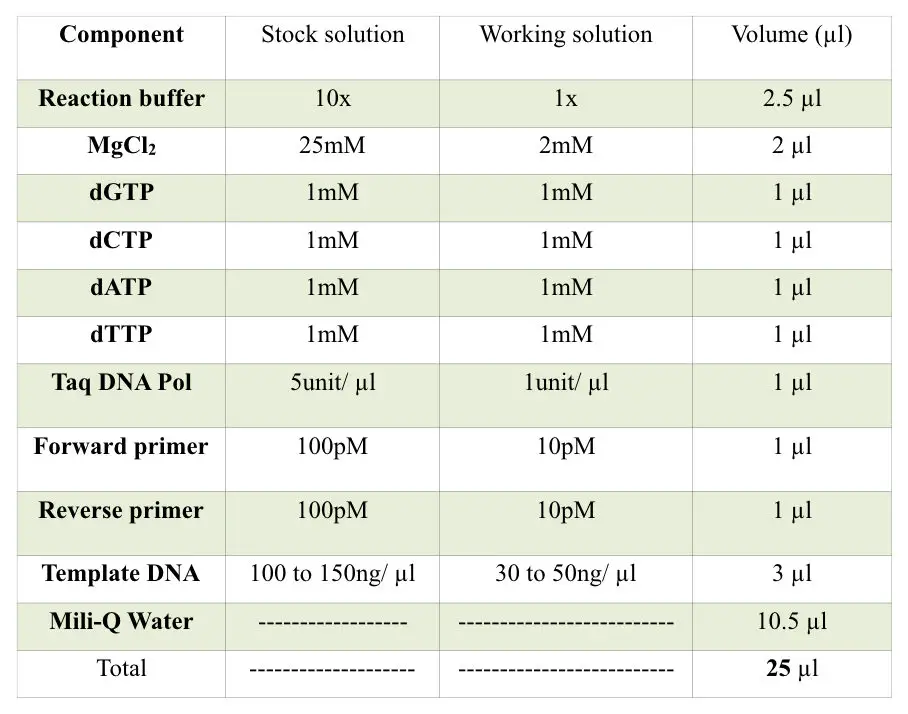
Preparation of 10X PCR reaction buffer:
| Chemical |
Concentration (10X) |
Concentration (1X) |
| Tris-HCl |
100mM (pH 8.0) |
10mM |
|
MgCl2 |
15mM |
1.5mM |
|
KCl |
500mM |
50mM |
|
Nonidet P40 |
1.0%W/V |
0.1%W/V |
|
Ammonium sulphate (NH4)2SO4 |
150mM |
15mM |
This recipe of the PCR reaction buffer is for 55% to 60% of GC content DNA. You can use this buffer directly or you can use one of the following chemicals as per your PCR need.
Conclusion:
Experience and expertise are required to play with the different components used in the PCR reaction buffer. Nonetheless, by doing the systematic and organize PCR protocol standardization experiments anyone can achieve expertise and excellent results.
Remember, never satisfied with your results because a combination of chemicals that you are using today that might not work in the future.


