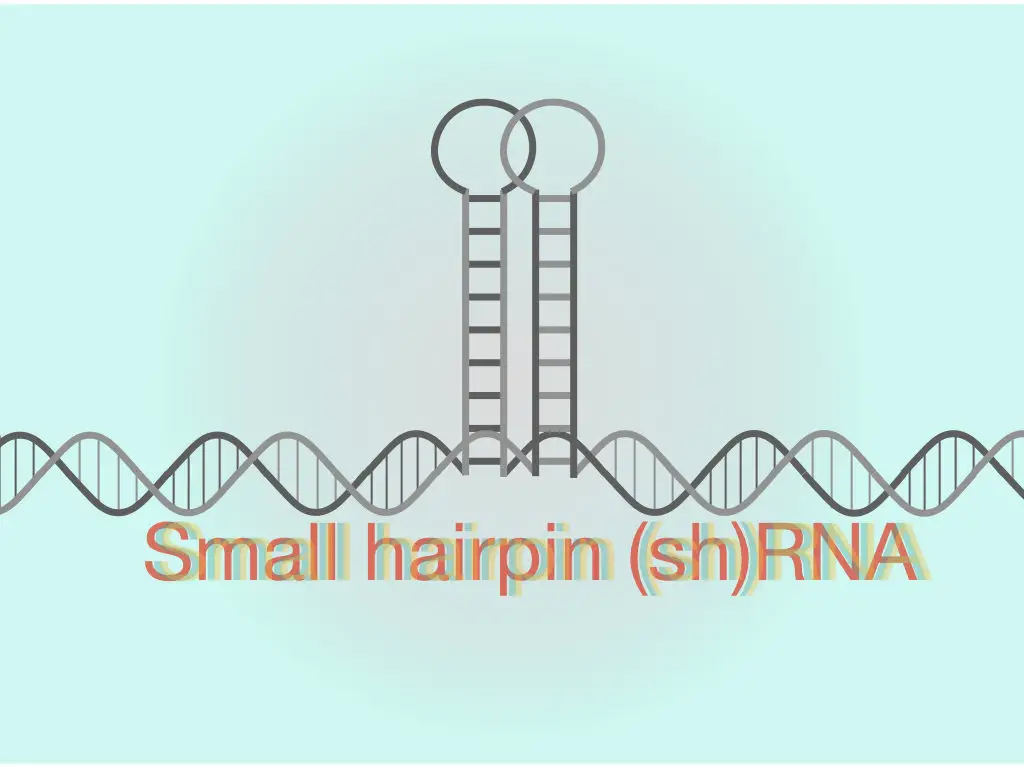
“The shRNA is an artificial dsRNA loop or hairpin applicable in the gene silencing studies which rely on the mechanism of RNA interference.”
The mechanism of shRNA action is similar to the siRNA. We had already discussed the siRNA. Read the article here: siRNA (Small Interfering RNA): Structure And Function
RNA isn’t a genetic material in us; it’s DNA but it helps to tailor various proteins. A few important, from many types of RNAs, are- rRNA, tRNA and mRNA. The ribosomal RNA and tRNA are a kind of helper molecules that favor the messenger RNA during translation, to make a useful protein.
Elaboratively, DNA replicates to make copies of genes, the mRNA forms from a gene through transcription (also known as a transcript) and a final protein product is formed by translation. This is the whole mechanism.
Single-stranded RNAs are the identity of a eukaryotic cell, which manufacture a protein using the gene or DNA template. Although, when a dsRNA is present in a cell, it is a sign of danger. While the dsRNA isn’t a genetic material of us, it must be destroyed.
dsRNA is possibly the exogenous pathogenic ribonucleic acid that will surely harm our cells. So that must be processed and removed. Using the mechanism known as RNA interference, cells destroy those enemies using it. The siRNA acronym as smaller interfering RNAs is much like the shRNA. siRNA helps in regulating gene expression.
The RNAi mechanism using either the shRNA or the siRNA can be applied for artificial gene silencing and gene expression studies. In a layman, we can say, shRNA and siRNA somehow similar, one is artificially created and one is a natural molecule.
In the present article, I will illustrate how the artificial dsRNA is used in the gene silencing technology, to cure disease. This article majorly focuses on shRNA and its various applications.
Related article: siRNA vs miRNA: 10 Major Differences.
| Abbreviation | Full name |
| mRNA | Messenger RNA |
| miRNA | microRNA |
| shRNA | Small hairpin RNA |
| siRNA | Small interfering RNA |
| RNAi | RNA interference |
| pri-miRNA | Primary microRNA |
| pre-miRNA | Precursor microRNA |
| RISC | RNA induced silencing complex |
| Ago | Argonaute protein |
Key Topics:
What is shRNA?
“The shRNA is our short hairpin RNA which is shorter, double-stranded ribonucleic acids, synthesized artificially and applicable in gene silencing experiments.”
Structure:
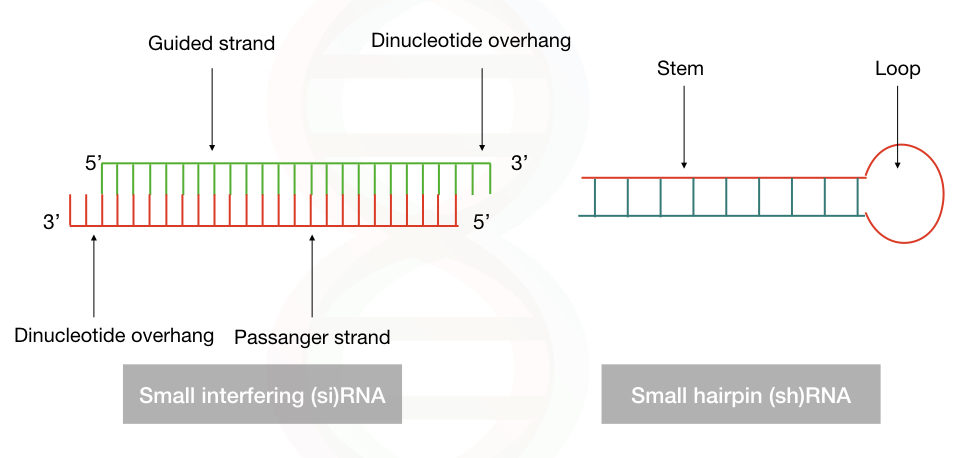
Often said as small hairpin RNA, the shRNA is a 20 to 25 bp polynucleotide chain of the RNA in which 4 to 11 nucleotides form a loop, a hairpin-like loop that binds to the mRNA molecule. Notedly, it lacks the dinucleotide overhang at the 3′ OH end, unlike the siRNA.
It provides a recognition site or binding motif for RNA-binding protein during the translocation and protects RNA degradation. It’s a secondary RNA structure loop that facilitates RNA folding.
Scientists use this double-stranded molecule for different purposes that we will discuss later on in this article. But before that, we need to know how the RISC and RNA interference mechanisms function.
If you don’t know either term, you can read the previous article here: Mechanism of RNA interference.
In a technical aspect, scientists use two types of shRNA- simple stem-loop (hairpin) RNA and microRNA-adapted shRNA. Both have different structures, compositions and functions.
A simple stem-loop RNA is made up of a 19 to 29 bp stem-loop region, 50 to 70 nucleotides long transcript and a dinucleotide overhang at 3’ end. It works like the pre-microRNA during RNA processing. This one uses the RNA pol III promoters.
Contrary, the microRNA-adapted shRNA is bigger in size than the simple one. It is as long as 250 nucleotides or more. Interestingly, it contains the native miRNA-like mismatches and that’s why it is more similar to the native miRNA.
It also has endogenous microRNA-like sequences on both flanking ends.
Indeed, the shRNA is a single-stranded RNA but by intermolecular base-pairing, it forms the double-stranded structure. The guided strand consists of exactly the same sequence as the target mRNA which is 21 to 29 nucleotides long. And an antisense strand that is correctly complementary to the guided strand. This interaction forms the ds-shRNA structure.
RISC (RNA interfering silencing complex):
The RNA interference silencing complex is formed by the alliance of dicer, siRNA or shRNA, argonaute (Ago) protein and dsRNA binding protein. Alike the siRNA, the shRNA also has the guided strand and the passenger strand.
Once the RISC finds the dsRNA, it recognizes the guided strand and localizes it on the complementary mRNA strand.
The ‘Ago’ protein present in the complex cuts the hybrid from the middle, followed by the subsequent endo and exonuclease cuts on the same mRNA.
The mRNA degrades into smaller pieces and can not undergo further translation. Hence the gene product which can be encoded by the mRNA can not forms. The short hairpin RNA can be artificially synthesized and used in vivo.
The expression of the shRNA can be induced using the plasmid, bacterial vector, or viral vectors. However, viral vectors aren’t recommended due to their infectivity.
If you understand the whole topic of RNA interference, one question may arise in your mind: why use the shRNA, when there is a siRNA already present in a cell?
Obviously, the shRNA is more effective than the siRNA because of the low mRNA degradation rate and turnover process.
Using a specially designed expression vector, the shRNA can be synthesized in vivo ( in a cell). Expression vectors are a great option to elevate the artificial shRNA-mediated process.
If you know how vectors work and manufactured, perhaps; you can understand how the process is done. In short, the expression vector must have a gene for the shRNA and the promotor sequence for robust and effective shRNA expression in a cell.
Technically, the RNA pol III or the DNA pol II promoters are generally utilized in shRNA-mediated RNA interference. The RNA pol III is more recommended because of having a higher shRNA expression rate.
In order to use the RNA pol III, the U6 and H1 promoters are commonly employed in the expression vectors for specific binding of the RNA pol III. unfortunately, promotes like CMV and EF1 aren’t so effective for shRNA expression.
(These all are the technical things! you just need to know about it don’t worry if you can’t understand!)
Once the expression vector is constructed, artificial techniques help penetrating it to the host or target genome at where the shRNA gene gets transcribed using the selected polymerase.
Read more on polymerase: Multifunctional DNA Polymerase: An Overview
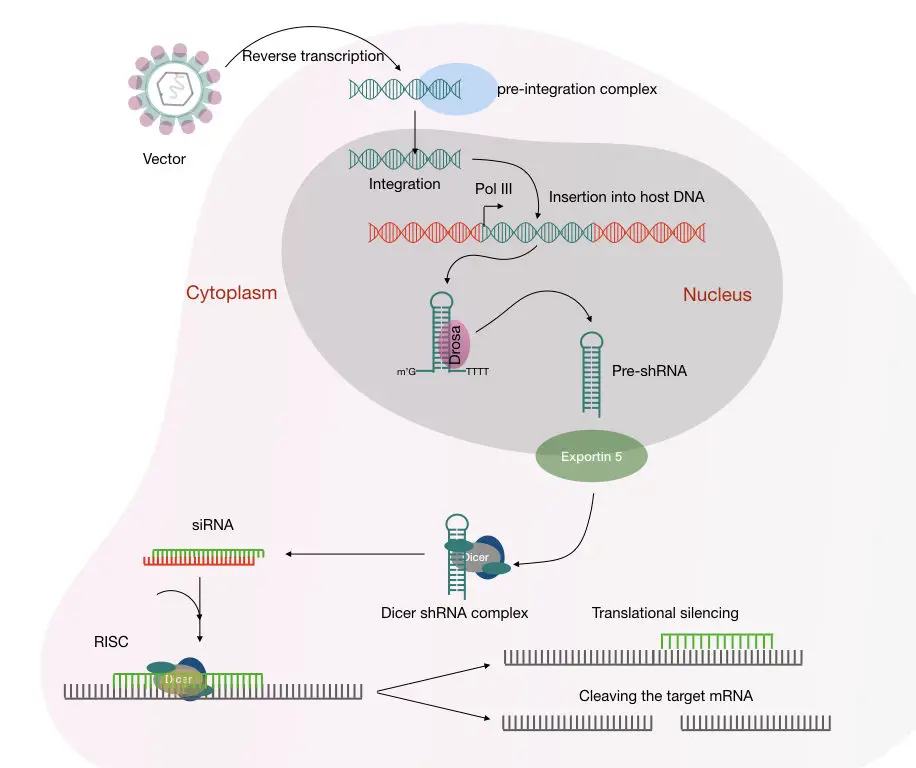
If we use the lentivirus-mediated transduction, the whole mechanism is like this:
shRNA is inserted into the cell and reverse transcribed into the DNA.
The ‘Drosha’ processes the pre-miRNA like pre-shRNA using the polymerase after the incorporation of the gene, gene promoter in the host genome.
Note: Drosa is a class 2 ribonuclease III enzyme involved in the dsRNA maturation.
Soon after, exportin5 protein transports the pre-shRNA to the cell cytoplasm. Here the pre-shRNA converts into mature shRNA. The dicer protein processes and cleave the siRNA-like dsRNA and loads it into the RISC.
Leaving the passenger strand behind the guided strand guides the complex to the complementary mRNA and binds to it.
The ‘Argo protein’, loaded in the RISC cleaves the mRNA and stops the protein formation governed by that mRNA. The mechanism of the shRNA mediated mRNA degradation is shown in the figure below,
Information:
In the year 2012, the first shRNA-mediated gene therapy for the Familial Adenomatous Polyposis was done using the gene knockdown method.
Read more: What is Gene Therapy? and How Does it Work?
shRNA mediated therapies:
Therapies like the shRNA are based on the DNA, and that’s why are known as DNA-based shRNA delivery therapies. DNA-based shRNA therapies rely on the use of the mechanism of reverse transcription. the DNA is formed and incorporated into the host genome.
Two common types are; plasmid-based transfection and selectable marker-based transfection.
The plasmid-based transfection technique has higher efficiency and transfection rate but at the same time, the use of retrovirus-like lentivirus makes its use restricted.
The selectable marker-based transfection is most recently developed and is effective too. In this technique, the selectable marker along with the shRNA gene and promoter sequence is incorporated to selectively eliminate those cells which aren’t transfected.
Selective marker-based transfection of shRNA helps to get a pure transfected culture or cell lines. The efficiency and transitivity of the shRNA depends on factors like,
- Vector selected for the therapy
- Selection and activity of the promoter selected
- Target cell types
- The artificial delivery system used
- The rate of epigenetic modifications
Besides, one of the biggest factors that decide the specificity and efficacy of the shRNA is how it is designed. To date, different companies have different algorithms and software to design the shRNA, computationally.
However, the best design should be achieved by taking into consideration the thermodynamic stability of the structure, formation of secondary RNA structure and position-dependent nucleotide preference.
So technically, the design of the shRNA should be powerful enough to fulfill these criteria;
- Higher rate of gene knockdown
- Can produce long-term and effective silencing to all cell types.
- The silencing effect should be stable in all cell types
- Can be processed accurately by the RNAi mechanism of a cell.
- Should have lower toxicity to cell
- Should have expression tracking to monitor the activity
- The molecule must be flexible enough to perform its activity in different cells.
Challenges in shRNA mediated therapies:
Let me tell you first that the mechanism or shRNA and how it actually works, isn’t clear to scientists yet. And therefore researchers face challenges to use it during gene silencing procedures.
The patient’s cells may develop a strong immune response against the shRNA.
Quite an understandable right! at first, the cell may treat our shRNA as the foreign, harmful and pathogenic dsDNA and induce a strong immune response to eliminate it.
It may be expressed uncontrolled.
Expression vectors have higher expressivity, meaning shRNA can be expressed in a much higher amount. cells can’t process it accurately.
Processing the wrong mRNA.
The whole process should be precise and target-specific. The shRNA only processes our target mRNA only. When it over-expresses, higher RISC saturation in a cell processes wrong mRNAs; mRNA other than the target one!
Overexpression can slip target-specificity which is yet another big trouble.
Off-target effect of shRNA.
Off-target is when our shRNA process the mRNA which isn’t the one we are interested in. As we said, the technology is new and not yet fully documented and understood therefore off-target effects are reported in some cases.
The therapy isn’t fully tested.
The therapy is not fully tested and might go off target, this can affects the expression of other genes as well.
Low transfectivity.
Meaning, the shRNA therapy has very low transfection efficiency. Specifically, non-dividing cells and other immune cells can’t be transfected precisely and effectively. It takes months to prepare shRNA-positive cells for transplantation.
Can’t work for all cell types.
Another disadvantage of this therapy is that it can’t fit all. Meaning, not all cell types are the target for the shRNA. it can work excellently for some cell lines, whine not for others.
Besides all these, one of the major limitations of shRNA-mediated RNA interference is the use of expression vectors.
Lentiviruses and other retroviruses are common vectors for shRNA. retrovirus are RNA viruses and aren’t good friends of ours. For example the HIV; we know how pathogenic it is. Henceforth, using retrovirus in therapy is a big risk factor.
Retrovirus may express its own genes and infect other cells. in short we can say, it’s a life-threatening decision to use retrovirus or related therapies for humans. In addition, the use of viral vectors can be harmful to the performer too.
Safety is one of the major concerns associated with the use of viral vector-mediated shRNA therapy.
Note:
As the siRNA and shRNA used the same mechanism of RNA interference, the choice of the therapy depends on the cell type, transient vs stable gene expression, time demand and the risk associated with it.
The general steps for shRNA mediated RNA interference given below,
Step1:
Selection of the target gene;
A minimum of two target sequences should be designed for one particular gene for increasing the efficiency of RNAi.
Step 2:
Construction of shRNA ranging from 19 to 27 bps complementary to the gene of interest, although the 19bp shRNA will give the best results.
Step 3:
In the next step, the shRNA expression vector is constructed either using the oligonucleotide-based cloning method or the PCR-based cloning method.
Note: PCR based cloning method is widely used because of the accuracy and the speed of PCR amplification. Read more on PCR: A Complete Guide of the Polymerase Chain Reaction
Here the single base mismatch can prevent hairpin formation thus it is very necessary to select the sequence having the best hairpin forming capacity.
Step 4:
In this step, RNA pol III or DNA pol II promoter sequences are selected for shRNA polymerization. Read our article on DNA polymerase: Multifunctional DNA Polymerase: An Overview
Note:
The lentivirus-mediated transduction is less toxic as compared to the AAS virus-mediated therapy, it can also infect the diving as well the non-diving cells efficiently which another benefit of using it.
Step 5:
After all that, the cells or the target cells are infected with the vectors having the shRNA for the gene knockdown study. The efficiency of infectivity is measured using quantitative PCR analysis.
Quantitative PCR or reverse transcription PCR is a tool used to measure gene expression. In addition, if the biological function of the target gene is known, its effect can be examined, whether the phenotype is suppressed or overexpressed.
The use of retrovirus isn’t a good idea and therefore the use of bacterial vectors is now in demand.
Read our articles on real-time PCR:
- Real-time PCR: Principle, Procedure, Advantages, Limitations and Applications.
- Reverse transcription PCR: Principle, Procedure, Applications, Advantages and Disadvantages.
Conclusion:
Researchers must find out the safest and secure therapy to alter gene expression. Perhaps, shRNA or siRNA-mediated therapies do not fulfill these criteria now, both methods have the potential to do well in the future.
shRNA-like RNA interference methods can be the best screening tool for cancer and related studies.
Sources:
- Moore CB, Guthrie EH, Huang MT, Taxman DJ. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol. 2010;629:141–158. doi:10.1007/978-1-60761-657-3_10.
- Principles of RNAi and shRNA design by Cellecta.