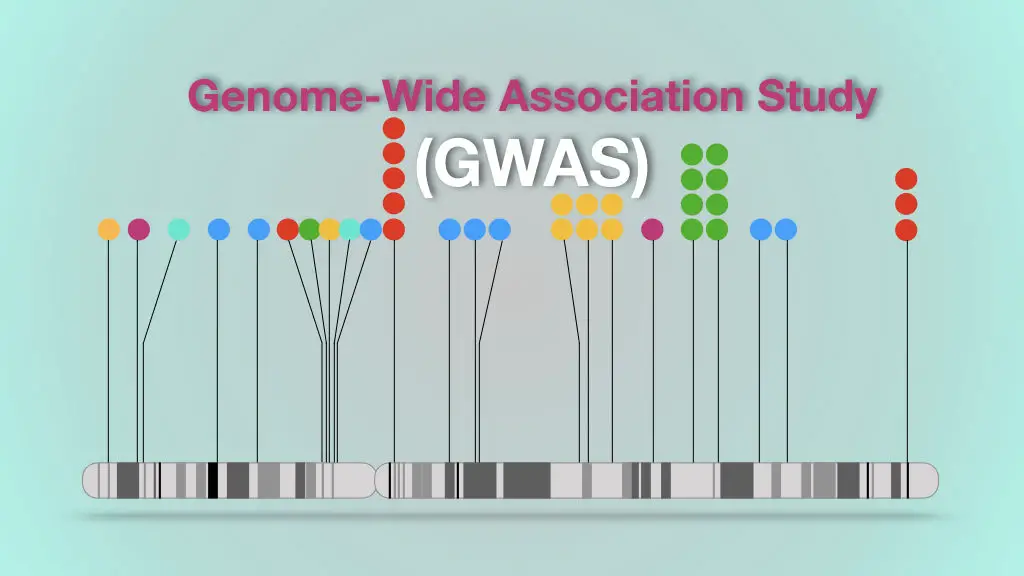
The Genome-wide association study is a method used to find out common SNPs associated with complex diseases.
For that, the single nucleotide variations from the whole genome are scanned and compared between case and control group.
Advanced genetic tools such as PCR, DNA sequencing and DNA microarray revolutionized the genetic diagnosis field.
These methods are so accurate and reliable, especially for the single gene defects. However, multifactorial and complex genetic abnormalities such as schizophrenia, diabetes or cancer are hard to screen using it.
A single gene defect occurs due to the mutation or group of mutations in one particular gene, for example, the beta-thalassemia.
The beta-thalassemia, an inherited blood abnormality is arisen due to the mutations in the beta-globin gene.
If we screen only a beta-globin gene, we can find out the mutation associated with the beta-thalassemia.
On the other side, so many genes and mutations are associated with complex disorders like cancer. Thus, when doing genetics for cancer, it is always a question for researchers from which gene to start or from where to start.
However, methods like the whole genome shotgun sequencing and next-generation sequencing can screen the entire genome of us. Read more on best genome sequencing methods: Three of the best genome sequencing methods.
The genome-wide association studies are the novel approach which detects the common genes or SNPs or alterations associated with the disease.
Although, that doesn’t mean that the mutations are itself responsible for the disease.
It simply means that the mutation (or more specifically the SNP) is associated with the disease.
In the present article, we will discuss the genome-wide association, the brief idea and its applications in genetics.
The content of the article is,
-
- What is a genome-wide association study?
- What is SNP?
- How the Genome-wide association studies carried out?
- Applications of Genome-wide association study
- Conclusion
Key Topics:
What is a genome-wide association study?
The genome-wide association study, often denoted as GWAS is an approach which scans many genomes at once, between the case and control for finding common genetic variations related to the complex disease.
The genome-wide association study is now used for the screening of complex genetic disorders such as Alzheimer’s, Parkinson’s disease, diabetes, cancer, Crohn’s disease etc.
It is a type of observational study based on the observation of phenotype and probability of its association with the genotype.
The first GWAS study was published in the year 2002 for myocardial infarction.
The second successful genome-wide association study was published in the year 2005, in AMD (Age-related macular degeneration) patients.
Interestingly, over the years, the method becomes so popular for investigation of complex diseases, scientists have now discovered over 2000 loci in the human genome associated with complex or multigenic disorders.
SNP chip for millions of different SNPs is nowadays available for the study. The SNPs, called the single nucleotide polymorphism is a marker used for the GWAS.
What is SNP?
A single nucleotide change or polymorphism occurred in a DNA sequence or in a gene or in a genome is called SNP, single nucleotide polymorphism.
The single nucleotide change might be an addition or deletion of a nucleotide. See the image below,
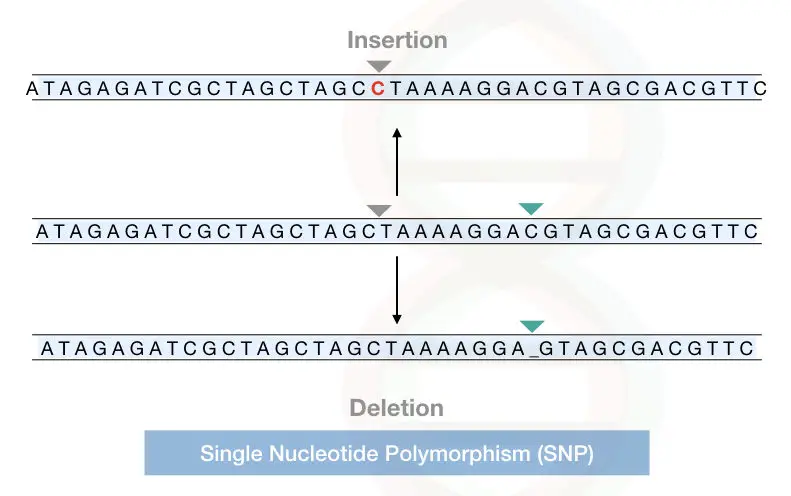
The SNP or single nucleotide alteration is very common in a genome, it may or may not be harmful or useful for the individual.
On an average, in a human genome, an SNP occurs at every 1000 nucleotides. As the data suggests, there are 3 billion base pairs are present into the human genome hence roughly 4 to 5 million SNPs are present in the human genome.
The SNP is a commonest alteration, often pronounced as “snips”. It occurs due to so many lifestyle reasons.
Studies suggest that depression and tension are one of the common reason for the occurrence of many SNPs.
Radiation, adverse food, adverse conditions, high-temperature, depression, tension, and other environmental factors are common causes of SNPs.
But as we stated above, the SNP may or may not be harmful to us, thanks to our DNA repair mechanism, the repair is done immediately.
Important information:
IVS 1-1, IVS 1-5 are the SNPs associated with the beta-thalassemia.
SNPs play an important role in the development of the disease. Again I am telling you that the SNP may be associated with the disease, not responsible for the disease.
Let’s take an example of diabetes,
Due to diabetes type 2, so many SNPs are originated in a genome (which are commonly found in the people with diabetes), by analysis those SNPs or identifying those SNPs or SNP we can predict the diabetic condition.
And that is the whole idea behind the GWAS.
[epcl_box type=”information”]“The SNP occurs due to environmental factors and lifestyle, not always strictly genetics.”[/epcl_box]
How genome-wide associated study carried out?
The genome-wide association studies are based on the case and control analysis. The study is a population-based study which includes a screening of many genomes at once.
The GWAS comprises of two components:
- The set of individuals with the disease
- The set (same type) of individuals without the disease
The whole blood or buccal swab is taken for the analysis and DNA is extracted using the ready to use DNA extraction kit.
Read more on DNA extraction: Different types of DNA extraction methods.
Using the SNP chip the genome of the selected candidates is analysed.
The SNP chip (as we discussed above) is constructed based upon the data of all the SNPs present commonly in a genome of us.
Another group of the individuals are the group of the same types of individuals as group one, but without the disease.
See the image below,
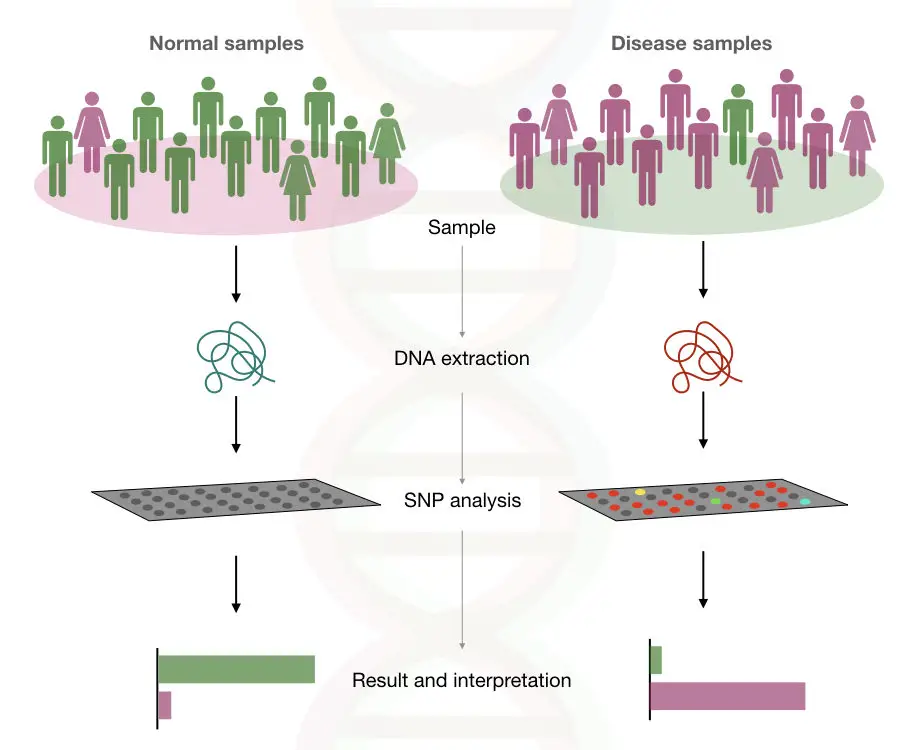
After the analysis of the SNPs of both the groups, the difference in the SNPs occurrence in both the genomes are called “associated with the particular disease”.
The SNP found in the diseased person or the frequency of the SNP found in the diseased person (as compared with the normal person) is predicted to be associated with the disease.
[epcl_box type=”success”]The SNP data of the disease persons or population is compared against the people without the disease.[/epcl_box]
Now let’s understand it by taking an example,
For instance, C to T mutation is present at location IVS 5 in a gene related to diabetes (assume that).
Now see the graph.
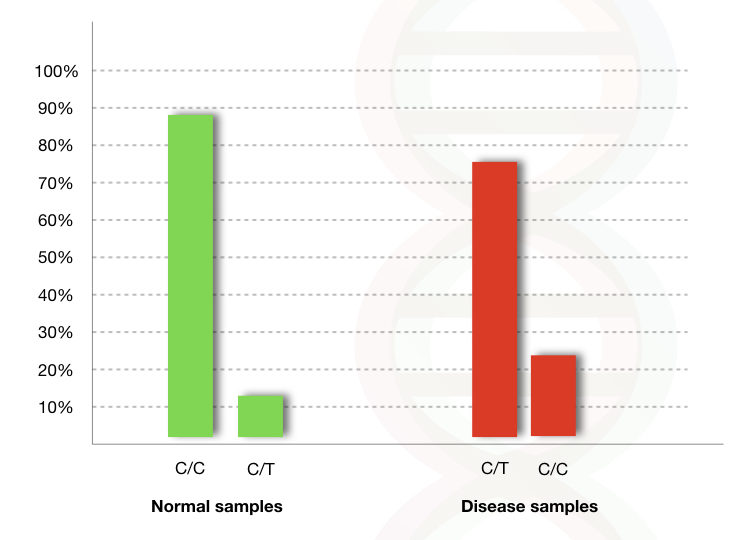
In the normal person, the C is present at IVS5 but in the diseased person, the T is present instead of C.
When we analysed 100 control and 100 disease person (diabetes), the C to T mutation (SNP) found in 76% of cases of diabetes, see the red bar.
The C to C is found in 88% of normal persons (see the green bar).
The results indicated that the C to T SNP is probably, strongly associated with the diabetic condition.
That is how the entire mechanism of genome-wide association study works.
In addition to this,
looking at the phenotype, a particular genotype related to that phenotype can be encountered through the GWAS.
Once the SNP or group of SNPs encountered, the scientists are looking after the genes on which those SNPs are located.
Then they do research on what protein and what type of protein that gene encodes, what is the function or role of it and what are the common mutations and other alterations in it.
By investigating all the data they will try to co-related that genes having the SNP with the disease conditions.
Also, the DNA sequencing technique used along with the GWAS helps in finding the cause of the disease by identifying the mutations associated with it.
Read more: DNA Sequencing: History, Steps, Methods, Applications And Limitations
TCF7L2, SLC30A8, IDE–KIF11–HHEX, CDKAL1, CDKN2A–CDKN2B, IGF2BP2, FTO, PPARG, KCNJ11, CAPN10. These are some of the genes associated with type 2 diabetes indicated through the genome-wide association study.
Remember!!
SNPs found in disease may directly not related to the disease but, it is commonly observed in those person having the disease.
Now see the graph below,
The graph is called a manhattan plot used for the GWAS study which shows different SNPs on different chromosomes.
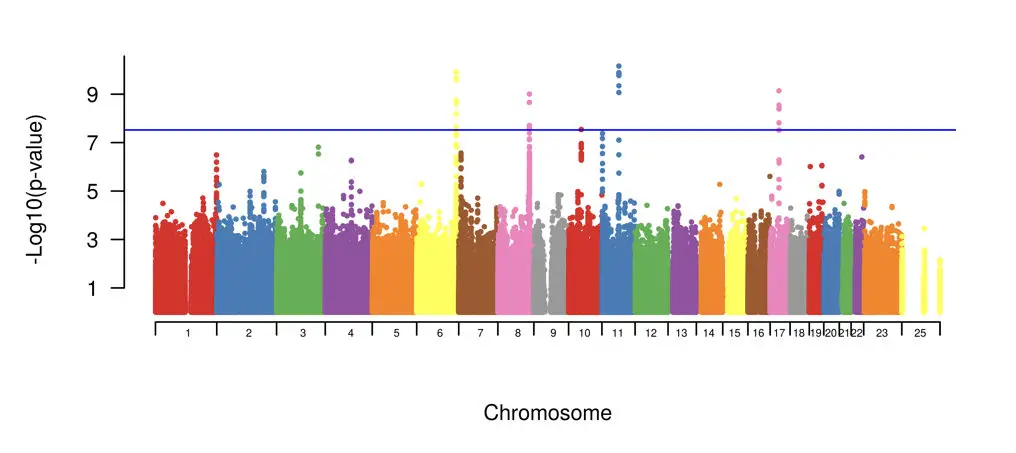
Applications of Genome-wide association study:
It is used in the study of complex and multigenic disorders such as diabetes type 2, Alzheimer’s, Parkinson’s disease, diabetes, cancer, Crohn’s disease, Rhegmatogenous retinal detachment, Diabetic retinopathy, Fuchs endothelial dystrophy, keratoconus, pseudoexfoliation, Age-related macular degeneration etc.
The genome-wide association study can also be used to encounter common phenotypic characteristics such as height, weight, body mass, BMI, blood pressure and insulin level etc.
It can also be used for the diagnosis of autoimmune disease and metabolic disorders.
Advantages of Genome-wide association study (GWAS):
- The genome-wide association study will help to find out the genes associated with particular complex disease.
- Though the genome-wide association study one can screen a large number of SNPs from the genome at once.
- The disease can be understood and treated well using GWAS data.
- It is further used in the development of personalized medicines associated with complex disease for a person.
- Hence personalized treatment can be possible.
- it can help in preventing complex genetic diseases.
The GWAS database:
The HapMap project started in 2005, is a storage of all the SNP data generated through the genome-wide associated studies worldwide.
All the data of GWAS are present online in NCBI’s “database of genotype and phenotype’, abbreviated as “dbGaP”
Related articles:
Conclusion:
Just within ten years, the genome-wide association study becomes one of the best personalized genetic screening tools.
It can be used in the prenatal screening for predicting the risk of the development of the particular complex disease.
By combining the tools like GWAS as well as DNA sequencing, one can screen the entire genome of an individual and can predict the chance of disease development in future.