“De novo sequencing is performed when no reference genome sequence is available to compare. Learn the concept of de novo genome sequencing in this article.”
Sequencing is a process of reading a nucleotide sequence, while whole genome sequencing is the same process but is applied to the genome level. Here, instead of a gene or sequence only, the entire genome of an organism is sequenced.
All the available DNA sequencing techniques have one common step- to compare it with the reference sequence or genome. They become only useful when reference sequence information or reference genome is available for comparison.
This will help investigate and study DNA or gene sequence and variants accurately.
However, without a reference genome, it is difficult to arrange or assemble the sequenced genomic regions in the correct order. The difficulty level increases with the repetitive regions and with gaps in the sequence.
De novo sequencing is a novel approach, a combination of wet lab design and dry lab compilation system to assemble or arrange the genomes of unknown origin or without reference information.
This article explains the overall concept, steps, procedure, advantages, limitations and applications of de novo genome sequencing.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy to understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Stay tuned.
Key Topics:
What is De Novo Sequencing?
De novo sequencing is an NGS (Next-generation sequencing)- based approach for reading unknown genomes when no reference genome or information is available. Technically, it has a slightly different experimental setup and a completely different bioinformatics environment for assembling.
The whole sequencing approach is designed in a way that makes scientists able to construct the genome of an organism from scratch. Still, both short and long-sequencing platforms support de novo sequencing.
Illumina, single and/or paired-end sequencing, single-molecule real-time sequencing and Nanopore sequencing offer de novo sequencing. Note that the Illumina platform works on short read sequencing.
On the contrary, long-read sequencing platforms (SMRT and NS) provide more accurate de novo sequencing results. The reason is that longer fragments are comparatively easy to assemble (fragments between >1K to 100K size).
In addition, the present approach has a more precise fragment assembly algorithm compared to the shorter read fragments.
Thus, the long-read sequencing technique is more suitable for de novo assembling, irrespective of its restricted sequencing capacity for repetitive and homopolymeric regions.
De novo sequencing, though, can be used to construct the genome of any organism but is extensively used in plant and microbial genome construction and reconstruction. We will discuss applications in a separate segment.
Steps and procedure:
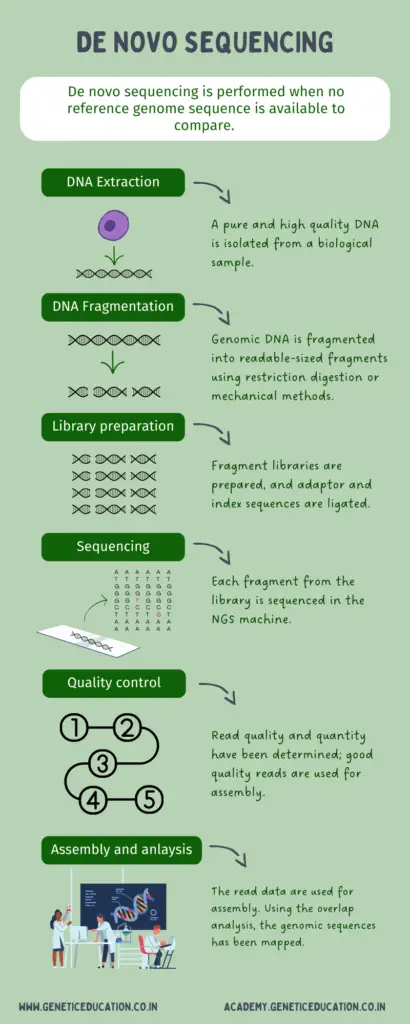
Much like the routine NGS experiments, the steps for de novo sequencing will almost remain the same. Here, I’m just providing the outline of steps.
DNA extraction: High-quality genomic DNA is extracted using the ready-to-use kit or automated DNA extraction unit from the target organism or sample.
Related article: Comparison Between Manual vs Spin Column vs Automated DNA Extraction.
Fragmentation: The unknown genome is then fragmented and used for library preparation. The fragments between 200 to 600 bp and >1k to 10K are generated for short read and long read sequencing, respectively.
(It depends on the platform you choose!)
Library preparation: Now the fragment library has been constructed, enriched and ligated with adaptors.
Related articles:
- Genomic DNA Library- Preparation and Applications.
- The Process of cDNA Synthesis and cDNA Library Preparation.
Sequencing: In the next step, the libraries are sequenced in the massive parallel sequencing approach. On a solid surface (chip or flow cell), the sequencing has been performed using the platform-specific chemistry.
Check out the table to learn about sequencing chemistry.
| Platform | Technique | Chemistry |
| Illumina | Short read sequencing | Sequencing by synthesis |
| Ion Torrent | Short read sequencing | Semiconductor |
| Oxford nanopore | Long read sequencing | SMRT |
| PacBio | Long read sequencing | SMRT |
*SMRT- Single Molecule Real Time Sequencing Technology.
Quality control: Quality control is a crucial metric in de novo sequencing. Bad quality and quantity readings result in poor assembly.
In the quality control process,
- Reads are first stored into FAST, FASTq, BAM or SAM file formats.
- Then, read numbers are considered for initial quality control purposes.
- After that, the read quality is determined.
- Next, the reads are trimmed, and adaptors and index sequences are removed.
- The computer algorithm combines all the key factors and determines the read quality.
- In the last step, trimmed reads are stored and used for de novo assembling.
Assembling and analysis: In the last step, all the fragments are arranged using the overlaps into the contings. Now, it is crucial to accurately determine the overlaps as we do not have any reference sequence to compare here.
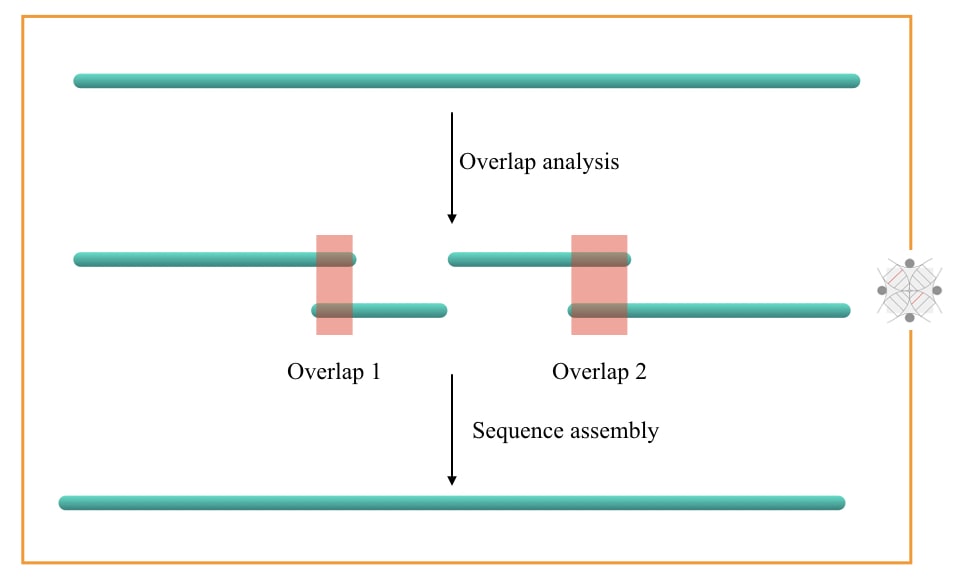
Specialized de novo genome assembling tools are used here. These tools are specially designed for de novo sequencing. Irrespective of the presence of gaps and “N” (undefined nucleotides) in the sequence, these tools can still perform read assembly.
Keep in mind that the important process in the de novo assembly generation is to prepare the contings by overlap read analysis. The experiment is designed in a manner that the overlap reads are generated. This provides ease in the assembly. Check out the above image.
Advantages:
One of the biggest advantages of the present technique is having the power to construct the genome sequence database of novel and unknown organisms. Scientists can discover and study the genomes of novel microbes and plant species using this approach.
Another crucial advantage is that it eliminates the bias linked to the reference genome.
De novo sequencing is significantly important for the species with no prior or least prior genomic information available.
Furthermore, like other NGS analyses, de novo sequencing allows scientists to identify and sturdy structural variants, novel mutations, new genes, and associated variants.
Disadvantages:
Despite its tremendous power to reveal novel genomes, de novo sequencing still has many critical limitations.
- First, the assembly can not be validated as no reference sequence information is available.
- Technically, it requires higher sequencing depth to assemble each read precisely.
- Keep in mind that the higher the sequencing depth, the higher the run time and, thus, the cost!
- It has higher error rates as we don’t have any reference data to correct or re-investigate the sequence.
- Complex, repetitive and homopolymeric regions pose additional challenges in the assembly process.
- It requires an extensive and costly computational and bioinformatics setup.
- Therefore, the de novo assembly technique is time-consuming, error-prone, costly and inefficient.
Applications:
Now, you may have a question.
When can we use it? Meaning, where can it be applied?
De novo sequencing is widely used in plant and microbial genetics and genome studies. Novel and unknown plant, microorganism and animal genome can be constructed from scratch.
Illumina’s unique de novo sequencing ecosystem was used to characterise the virus genome of nCoV (coronavirus) and helped during the COVID-19 pandemic.
Researchers at ARC of South Africa used the Illumina sequencing platform for the de novo sequencing of sweet potato viruses. The identification and mapping helped to improve food security.
Ancient samples and extinct species’ genomes without prior sequence data can also be sequenced. This will give us valuable genetic information from those species or ancient samples.
In addition, through the study of these ancient samples, scientists can decode evolutionary history.
It is also a valuable tool in outbreak and pandemic studies, antibiotic resistance investigation and vaccine development.
Related article: What is Resequencing, and Why and When is it Needed?
Wrapping up:
De novo sequencing is underrated but the most valuable technique, particularly for microbial study. Approaches like long-read and real-time sequencing make the de novo sequencing process effective. Still, it has some crucial limitations.
Repetitive sequence nature, gaps and unavailability of reference genome information are the major challenges in the present process. I hope you like this article. Do share it and subscribe to Genetic Education.
If you want to strengthen your sequencing knowledge, you can participate in our Next- Generation Sequencing mastery course. All the course information is provided on the course page. Click the link and inquire now!
Resources:
Camilli A. De Novo Genome Sequencing, Annotation, and Taxonomy of Unknown Bacteria. Cold Spring Harb Protoc. 2023 Jan 3;2023(1):1-3. doi: 10.1101/pdb.top107847. PMID: 36283838; PMCID: PMC10586727.De novo sequencing by Illumina.


