“Parameters that influence the Sanger sequencing results are good quality extract, amplicon purification, primer designing and managing leftover ddNPTs. These parameters can improve the Sanger Sequencing results, certainly.”
Let’s findout how to deal with each parameter individually to create a combined effect on the overall Sanger results.
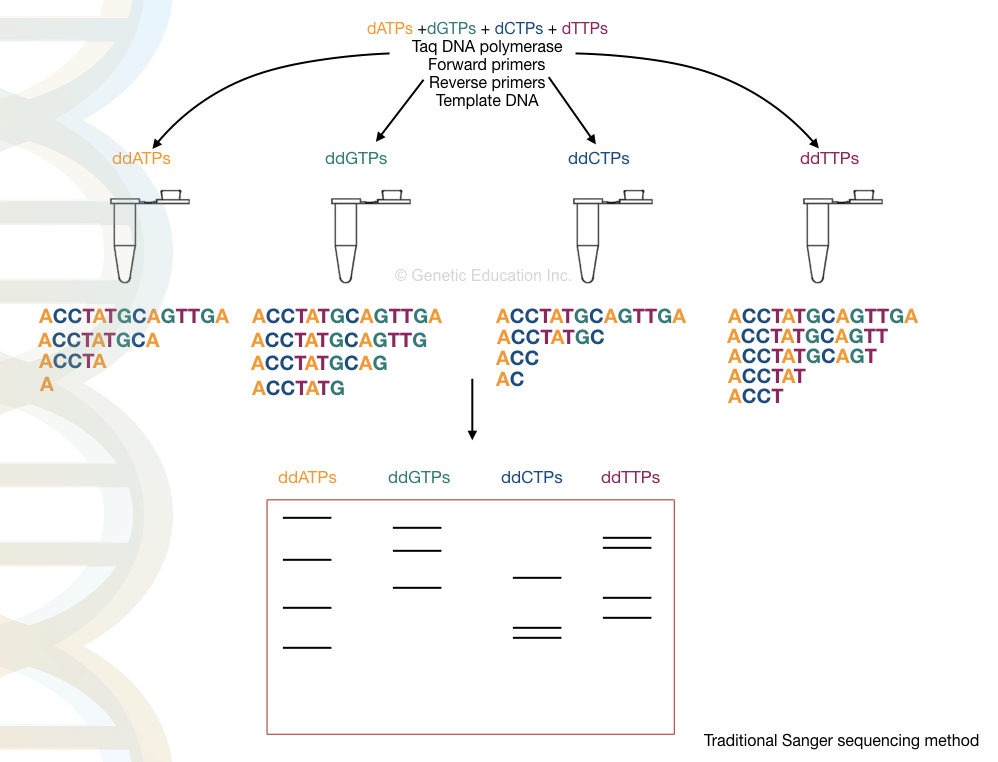
Many different types of DNA sequencing are popularly known but Sanger sequencing is by far the most crucial discovery much like the PCR. Discovered by Sanger, two important factors Sanger sequence has are the use ddNPTs and synthesis termination.
Four different reactions are set up with a standard reaction mixture. In addition, four different ddNTPs are added. As ddNTPs lack the hydroxyl group, whenever the polymerase ads ddNTP, the synthesis terminates which is also referred to as chain termination.
After completion of the reaction, the amplicons are run on a PAGE gel and each fragment is analyzed to generate a sequence, manually. An automated system is now available that sequences and runs the sample simultaneously. This approach is more speedy, easy, cost-effective and reliable.
The automated Sanger sequencing platform is now popular and employed for research and diagnostics too. Although the entire system seems accurate and perfect, extrinsic factors like DNA extract, primer designing, amplicon purification, removal of ddNTPs and pipetting and common lab expertise can influence the results, adversely.
At the same time, by taking care of such factors will boost the performance too. So in this article, I will give you 5 tips using which you can surely improve your sequencing results.
Note that I am assuming that you know the Sanger sequencing well. So let’s start.
Key Topics:
How to improve Sanger sequencing results:
Here are my five tips or you can say optimization options that will improve your Sanger sequencing results.
1. Quality and quantity of DNA:
“I only go with DNA having good quality and quantity. And I think that certainly is so important too.”
Debayan Baidya. Research fellow, CHARUSAT university, India.
Quantity and quantity of DNA extract are so crucial to get good downstream results. It’s a super easy task but one has to have a decent working hand in extraction. Experts advise using manually extracted DNA over ready-to-use kits.
Manual DNA extraction protocols are controllable, we can optimize them as per the requirement. We can improve the purity and yield of DNA and once the protocol is set, we can use it, repeatedly. In fact, manual protocols yield more DNA.
Importantly, I do not advise using manual DNA extraction for a smaller volume, precious and important biological sample. Take an expert’s advice in that case. The DNA should have ~1.8 purity (260/280 ratio) and 100- 300 ng concentration, Said Baidya D further.
You can read our series of articles to learn more about DNA extraction: DNA extraction.
Consequences of the bad quality extract are poor quality sequence reads, high background signals and reaction failure. Even, I recommend using optimized phenol, chloroform and isoamyl alcohol DNA extraction protocol.
2. PCR amplicon purification:
“My advice is to take amplicon PCR seriously. It boosts the quality of reads during analysis.”
Tushar Kachhadiya. Geneticist, SN gene Lab, Surat, India.
PCR amplicons of desired fragments should be pre-purified before sending for sequencing. There are a couple of reasons for that. The leftover single-stranded sequences, unused primers, primer-dimers and free nucleotides will create problems in amplification and consequence in poor quality reads.
To avoid it, purify it first. Two common techniques are spin-column-based purification and Exonuclease-based purification. Particularly, for sequencing, ExoSAP-IT PCR product cleanup reagent is used. I advise using this and not the spin-column technique, Kachhadiya T said.
The enzyme here digests the unused nucleotides, primers and single-stranded DNA fragments or leftovers. Along with this, it protects our precious PCR amplicons too. The purification technique is super easy, add the enzyme to a tube, incubate it, heat it and the sample is ready.
Note that the manufacturer’s guide has the protocol to use reagents. Refer to it before use.
3. Primers for sequencing:
“Expand the primer annealing site at least 50 nucleotides on both sides to avoid trimming of the desired sequence.”
Dr Jigar Suthar, Genetic technologist, Unipath Lab, India.
First, let me explain that the primer design process for sequencing is similar to PCR. Again the process is easy to perform but one thing beginners have to keep in mind.
Very short PCR product does not migrate properly. Creates poor quality traces or reads at the beginning. Further to this, the end of sequencing is also not good because of the same reason, the longer products can’t separate effectively in capillary electrophoresis.
So it is advised to design primers that bind 50-60 nucleotides before and after our actual amplicon. For example, if your product is 200bp then design primers of 320bp (60 above the 200bp fragment and 60 below the fragment).
During the analysis of results, we can trim such unwanted and poor-quality reads to generate excellent sequencing results.
4. Purification of sequencing product before analysis:
Kachhadiya T is a sequencing expert at SN gene lab, Surat, India and shared his practical knowledge further by adding more value to this topic.
“The most crucial step in Sanger sequencing for improving the quality of results is product purification. Although, it’s not a tedious process but is important to understand what problems it can cause.”
The unused ddNPTs cause a ‘dye-blob.’ The aggregation of ddNPTs in the reaction produces unwanted signals. Such signals produce unwanted peaks which should be manually assessed.
Sequencing product cleanups are available thankfully that can overcome dye-blob mostly. Still, varying degree of dye-blob in the electropherogram is an indication of inefficient and poor quality sequencing reads.
Learn more: Sequencing reads.
5. Accurate pipetting:
This advice is from my side.
DNA extraction, amplicon purification, and sequencing cleanup all are important steps but highly depend on the pipetting hand of lab personnel. Meaning, the effectiveness and accuracy of sequencing highly depend on how you prepare and perform pipetting during each step.
Each reagent should be weighed correctly in not only sequencing but also in all molecular genetic experiments. If you don’t have good pipetting skills, learn it, practice it and then use it.
Trust me pipetting with the best hands will surely improve your results. You will notice improvements. In addition, it is also important to use calibrated and good-quality pipettes for such important experiments.
Read more on Sanger sequencing:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- Detailed History of DNA Sequencing
- Why Should Life Science Students Learn Sanger Sequencing? – Top 6 Reasons
Wrapping up:
I left my research career very early but gained substantial experience in various techniques. These are some of the tips from experts and my side that can improve your sequencing performance. Even if you are new, you can implement it.
Sanger sequencing, as per my knowledge is a well-executed and excellent sequencing platform by far, undoubtedly. Recent automation and robust approach have revolutionized the entire field of research and diagnostics.
I hope this article will add more value to your sequencing knowledge.


