“A recent research paper published in Nucleic Acids Research uncovered the role of the cohesin protein complex in gene regulation.”
Key Topics:
Summary
- Gene expression is defined as the number of gene copies that undergo transcription and are translated into proteins.
- Epigenetics is the study of changes in gene expression, rather than changes in the nucleotide sequence.
- Cohesin is a protein complex, majorly participates in the DNA looping process.
- Abnormal cohesin activities lead to conditions like cohesinopathies.
Research insights:
Fettweis et al. published a recent study in Nucleic Acids Research entitled “transcriptional factors form a tertiary complex with NIPBL/MAU2 to localize cohesin to enhancer,” which showed the significance of a tertiary complex in localizing the cohesin protein on the chromatin.
The tertiary complex that they mentioned in their research, is a combination of NIPBL, which is a cohesin loading complex, MAU2– a partner protein and glucocorticoid receptor (GR) as a transcriptional factor.
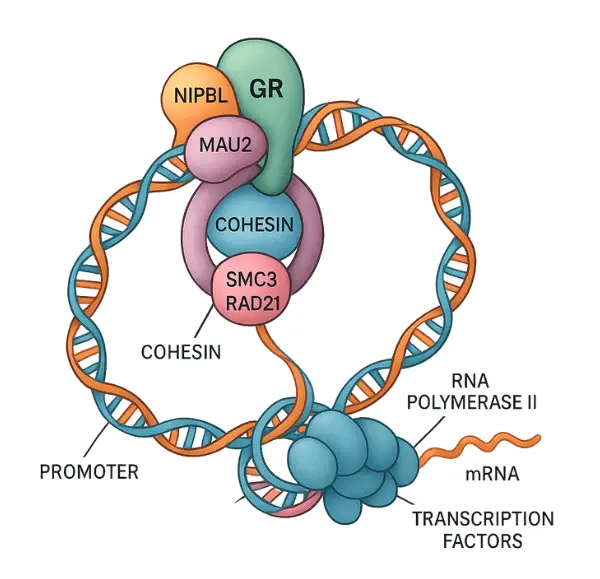
The combined action of all three factors directs cohesin to a specific region of the genome, known as an enhancer. This enhancer functions as part of the gene’s regulatory toolkit, enabling the binding of transcription factors to initiate gene expression.
A gene needs a promoter to initiate transcription, though, cohesin, the loop-forming protein, when guided by the tertiary complex, makes a loop and provides the close contact between the gene and its promoter.
Here, the MIPBL-MAU2-GR complex plays a crucial role as it precisely places the cohesin protein on the target gene and allows targeted gene expression. This finding showed how important the cohesin-mediated loop formation is for gene regulation.
Further investigations demonstrated that any abnormality in the present complex leads to a specific genetic condition, known as ‘cohesinopathies.’
What’s cohesin do?
Cohesin is a specialized protein that participates in chromatin regulation. The multicomplex protein, cohesin, has many subunits. The list is given below.
| Subunit | Function |
| SMC1 | Structural Maintenance of Chromosomes protein 1 – forms a coiled-coil structure |
| SMC3 | Structural Maintenance of Chromosomes protein 3 – pairs with SMC1 |
| RAD21 (also called SCC1) | Acts as a “bridge” connecting SMC1 and SMC3 |
| SA1/SA2 (also called STAG1/2) | Regulatory subunits that modulate binding and release |
The primary function of cohesin is to correctly segregate DNA after replication. In addition, it helps in loop-mediated gene regulation and DNA repair.
The present study uncovered the complexity of the cohesin-mediated gene regulation pathway, using techniques like ChIP-seq, co-precipitation, RNA interference knockdown, quantitative real-time PCR, luciferase reporter assay, and atomic force microscopy.
The key takeaways from the present findings are explained here.
Key takeaways
- The present research identified the novel tertiary complex (NIPBL-MAU2-GR), having an extensive role in targeting the cohesin protein to a gene enhancer.
- This depicts that cohesin is not involved in random DNA binding and looping. The tertiary complex assesses binding at a specific and target location on a gene enhancer.
- This special arrangement activates a specific gene to undergo transcription.
In conclusion, this novel finding enhances our understanding of cohesin-mediated DNA loop formation and reveals how a multi-protein complex precisely targets specific genes for expression.
Source:
Fettweis, G., Wagh, K., Stavreva, D. A., Jiménez-Panizo, A., Kim, S., Lion, M., Alegre-Martí, A., Rinaldi, L., Johnson, T. A., Gilson, E., Krishnamurthy, M., Wang, L., Ball, D. A., Karpova, T. S., Upadhyaya, A., Didier Vertommen, Recio, J. F., Estébanez-Perpiñá, E., Franck Dequiedt, & Hager, G. L. (2025). Transcription factors form a ternary complex with NIPBL/MAU2 to localize cohesin at enhancers. Nucleic Acids Research, 53(9). https://doi.org/10.1093/nar/gkaf415.


I admire the passion behind this post. Keep up the great work!