“Sequencing by synthesis is Illumina’s unique chemistry behind their NGS platforms. Gain a basic understanding of SBS and its principle and chemistry in this article.
Illumina is a pioneer in sequencing technology; NGS (Next-generation sequencing), in particular. Each NGS can do whole-genome, exome, transcriptome and metagenomic analysis.
Sequencing by synthesis is a unique chemistry used in the Illumina NGS machines. It is different from the conventional and most popular Sanger sequencing chemistry. It’s more valuable and advanced than the Sanger chain termination method.
In this article, we are going to discuss the sequencing by synthesis (SBS) approach used in NGS for sequencing. We will understand the concept, mechanism and entire chemistry, benefits and limitations.
This is another installment of our sequencing learning class. Stay tuned.
Related article: DNA Sequencing: History, Steps, Methods, Applications and Limitations.
Key Topics:
What is SBS?
SBS is a sequencing-by-synthesis technique for sequencing DNA and RNA. It was developed by Illumina and used in their NGS platforms. The term literally means that sequencing occurs during the synthesis process.
This means it occurs during DNA replication, in vitro. Put simply, when the polymerase synthesizes the target nucleic acid strand, the reading or sequencing process co-occurs.
Each nucleotide is added by the polymerase and read by the unique chemistry and instrumental setup used. Let’s discuss the chemistry and principles behind SBS.
Principle
DNA synthesis led by the polymerase adds labeled nucleotides during each round of sequencing. Each nucleotide is added to its complementary part, read (imaged), recorded and enzymatically degraded to prepare the template to add the next nucleotide.
As each nucleotide consists of a unique color label, the captured image is used to translate the read signals into nucleotide sequences. In NGS, this SBS occurs in a massively parallel fashion millions of times.
Concept and Chemistry
The concept of sequencing by synthesis is not new, attempts were made during 1988. Previous attempts failed due to the activity of the polymerase that disallows modifying either the template strand or nucleotide being added.
Chemically cleavable fluorescent nucleotide analogs, used in recent chemistry, have been developed as a successful option for NGS. Now, let’s understand the concept comprehensively.
Extensive NGS process explanation is excluded from the present article, only the sequencing part is included. As we know, the forward and the reverse templates are read separately during Illumina paired-end NGS sequencing, first, the reverse template DNA is washed off and only a forward strand will remain.
Also, note that cluster generation has also been completed during bridge amplification, now we are on the sequencing step. Using the forward template, the first sequencing primer comes into action to produce the first read.
Fluorescently tagged nucleotides are added, one in each cycle to their complementary nucleotide on the forward strand. All differently color-tagged nucleotides are added in this manner. Polymerase looks after the correction addition of nucleotides to their complementary part.
After completion of the synthesis part, the light source is passed through each nucleotide and emits fluorescence. Using 4-color chemistry four different fluorescent lights have been recorded during the sequencing-by-synthesis process.
Now, it is interesting to know that polymerase can’t allow the incorporation of a labeled nucleotide (whose chemical structure is altered). So once, the labeled nucleotide is inserted and fluorescence is recorded, the polymerase removes the fluorescent tag and makes the sequence ready for another round of synthesis.
Once the signal is recorded the fluorochrome is washed off to make the sequence ready for another round of sequencing. This is the broad chemistry behind sequencing by synthesis. Now coming to the imaging part.
Illumina has various imaging chemistries. The 4-color SBS channel chemistry is the oldest one, while the 2-color and single-color channel chemistry is the latest one.
4-Channel SBS Chemistry
Here, four differently labeled nucleotides are added to the flow cell. Polymerase incorporates each nucleotide based on sequence complementation. Four different wavelength bands are generated by capturing four different images. The image analysis software processes the image and generates the sequence.
4 different fluorochromes, 4 different cycles and 4 different base calls.
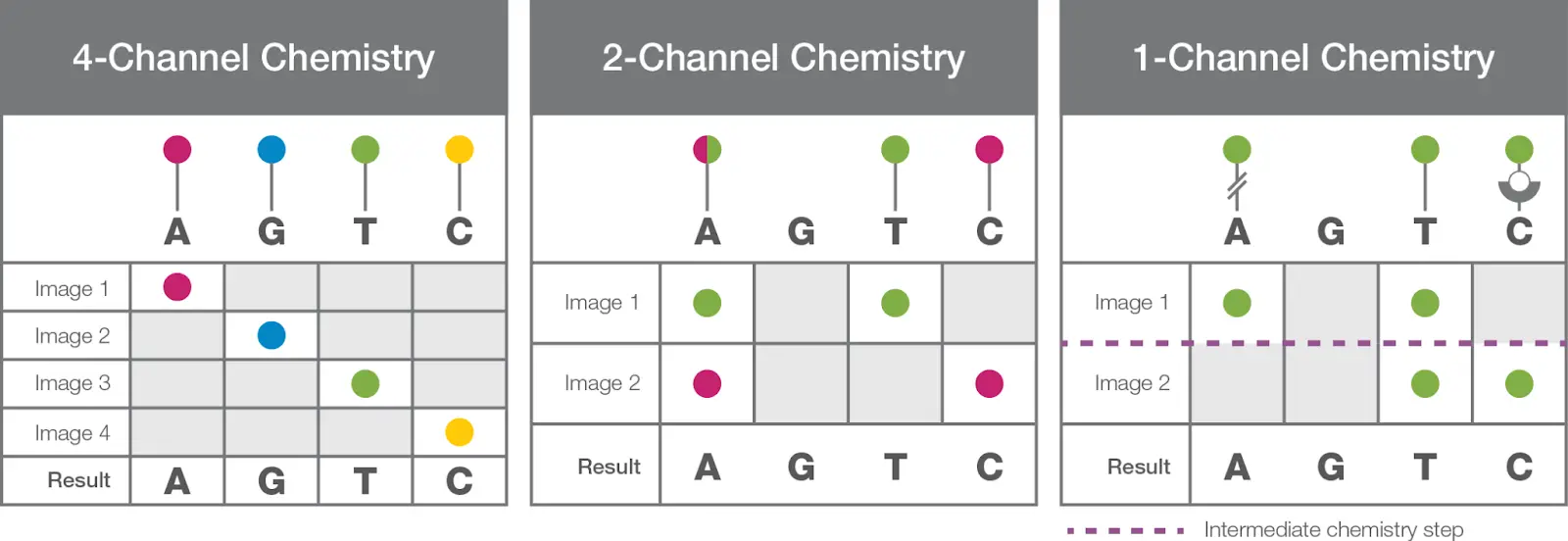
2-Channel SBS Chemistry
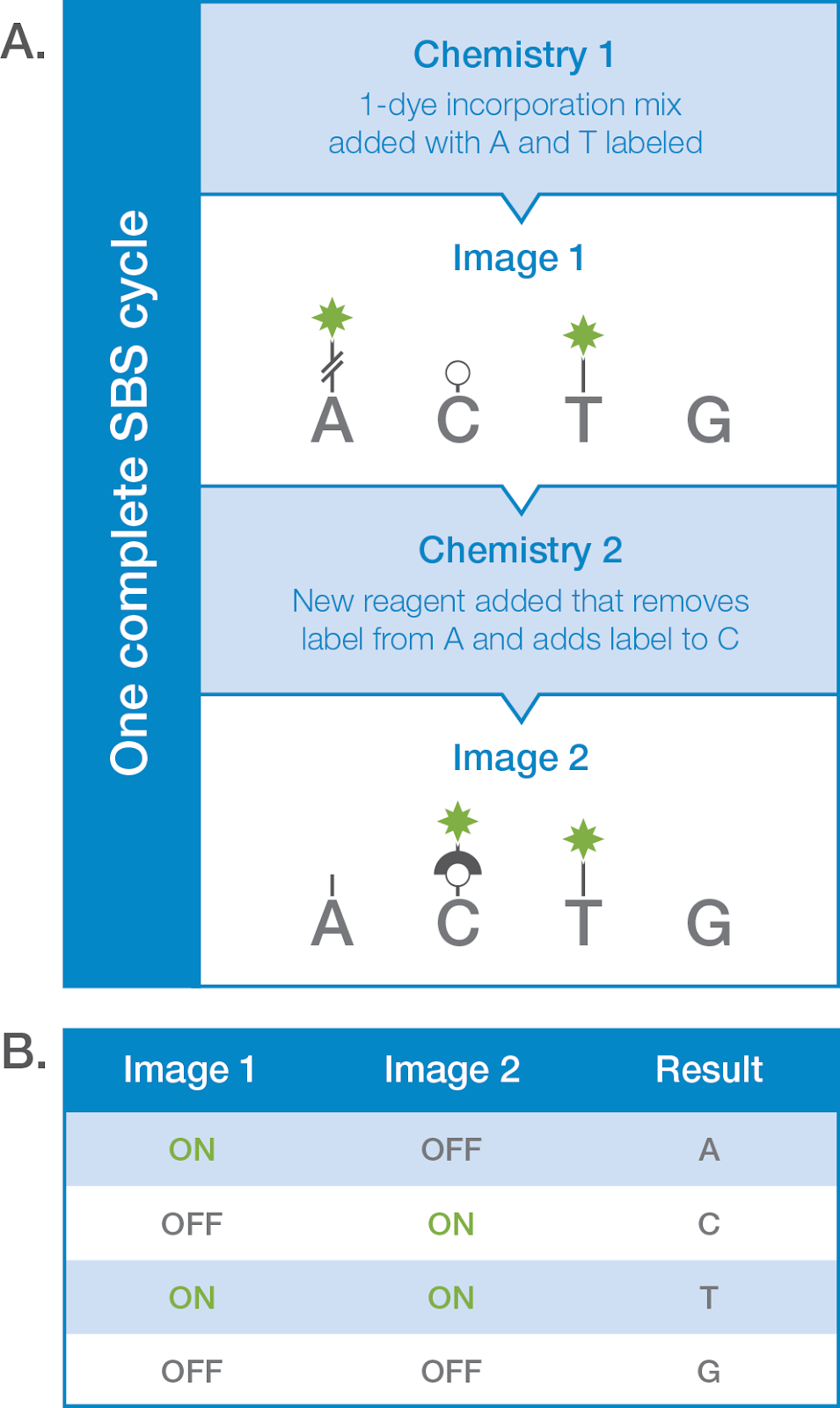
Now, in this chemistry, only two separate fluorescent dyes are used and thus, only two cycles are performed during sequencing. An additional red and green filter is incorporated to process all four images.
Here, thymines are labeled with green, cytosines with red, adenines with a combination of red and green and guanines as dark (permanent). 2-channel SBS saves time, cost and reagents along with increased accurate reading.
Single-channel SBS Chemistry
One-channel SBS uses only a single fluorochrome to sequence all four nucleotides. Here, sequencing occurs in nanowalls in which each cluster is directly attached to each photodiode or pixel. Only two images are captured to read all four nucleotides.
| 4-channel | 2-Channel | 1-Channel | |
| Fluorochrome | 4 | 2 | 1 |
| Cycles | 4 | 2 | 1 |
| Image captured | 4 | 2 | 2 |
| Platforms | MiSeq and HiSeq | MiniSeq, NextSeq and NovaSeq | iSeq100 |
Sanger sequencing vs SBS
Previous sequencing platforms such as Sanger sequencing and pyrosequencing have several serious limitations. Let’s take an example of Sanger sequencing.
Sanger sequencing works on the principle of chain termination method which uses ddNTPs along with dNTPs in the reaction, while the SBS works on the principle of synthesis only, and uses labeled dNTPs only.
Sanger sequencing requires gel preparation and gel-based analysis, which is inefficient and error-prone, while the SBS doesn’t need any gel preparation or gel-based analysis. It uses a unique chemistry and computation program for results analysis.
Sanger sequencing needs a larger sample volume while SBS needs a small amount of starting material. In SBS, sequence enrichment occurs through bridge amplification.
Sanger sequencing can read a single fragment in a single reaction while sequencing-by-synthesis can sequence millions of DNA fragments in a massively parallel reaction.
Hence, Sanger sequencing is a limited throughput technique while SBS-based NGS is a high throughput technique.
In addition, homopolymer and highly repetitive regions are difficult to sequence accurately, however, SBS chemistry has a unique characteristic of natural competition among all four nucleotides that allows the incorporation of exactly correct nucleotides.
Related article:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- What is NGS?- Definition, Principle, Steps, Chemistries, Advantages and Limitations
- De Novo Sequencing: Steps, Procedure, Advantages, Limitations and Applications
Wrapping up
The unique chemistry of Sequencing-by-synthesis has uplifted the performance of next-generation sequencing many folds. It’s speedy, accurate, precise, high-throughput and fully robust.
Therefore, SBS has been extensively employed for cancer research. It can accurately study various copy number variations, SNPs, rare variants and repetitive genomic regions.
At present, Illumina, Roche, Helicos, Pacific Biosciences and Life technologies are also using sequencing-by-synthesis chemistry in their various NGS platforms. I hope this article helps you learn about the sequencing by synthesis process and will help you choose an assay and machine for your experiment.
Sources:
Explore Illumina sequencing technology by Illumina.
Guo J, Yu L, Turro NJ, Ju J. An integrated system for DNA sequencing by synthesis using novel nucleotide analogues. Acc Chem Res. 2010 Apr 20;43(4):551-63. doi: 10.1021/ar900255c.
Ju, Jingyue, Dae H. Kim, Lanrong Bi, Qinglin Meng, Xiaopeng Bai, Zengmin Li, Xiaoxu Li et al. “Four-color DNA Sequencing by Synthesis Using Cleavable Fluorescent Nucleotide Reversible Terminators.” Proceedings of the National Academy of Sciences 103, no. 52 (2006): 19635-19640. Accessed January 23, 2024. https://doi.org/10.1073/pnas.0609513103.
Fuller CW, Middendorf LR, Benner SA, Church GM, Harris T, Huang X, Jovanovich SB, Nelson JR, Schloss JA, Schwartz DC, Vezenov DV. The challenges of sequencing by synthesis. Nat Biotechnol. 2009 Nov;27(11):1013-23. doi: 10.1038/nbt.1585.


