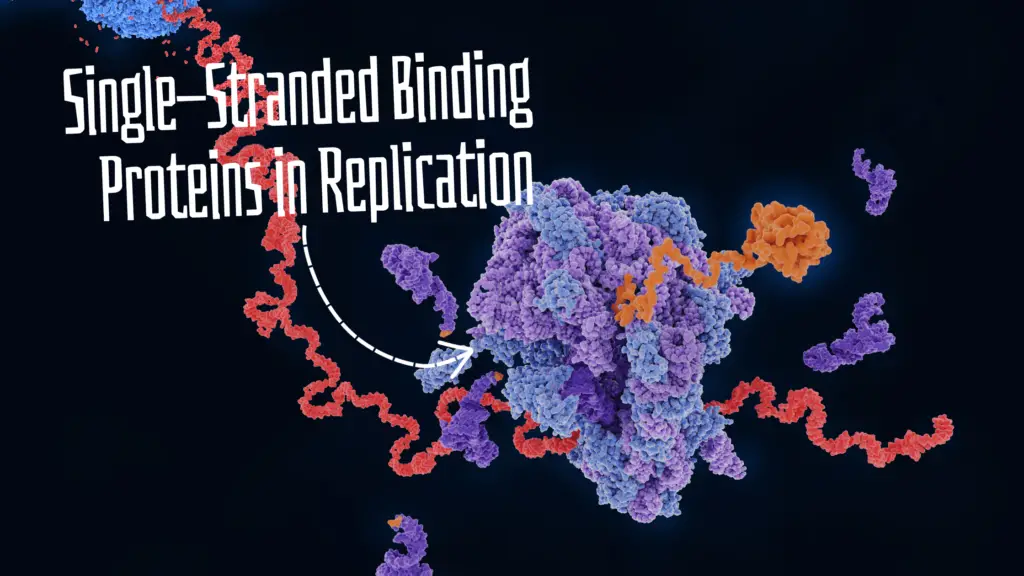
“Single-stranded binding Proteins are multi-domain DNA proteins and help during replication to stabilize the single-stranded DNA. Learn about the SSB protein in this article.”
The general process of DNA replication includes three steps— Initiation, Elongation and Termination and many different enzymes and proteins. It works to prepare an exactly similar DNA copy.
Double-stranded DNA is a highly stable structure. However, the collective efforts of helicase and topoisomerase make available the single-stranded DNA form for replication.
However, the thermodynamically unstable ssDNA hampers replication. SSB proteins remain attached to the single-stranded DNA and allow effective replication. That makes them also an important ‘game player’ for replication.
In previous articles of this series, we discussed helicase, topoisomerase, ligase, polymerase and primase. Now, in this article, I will explain the structure, function, mechanism of action and various types of SSB proteins used in DNA replication.
Disclaimer: Information provided here is collected from peer-reviewed resources and re-presented in an understandable language. All the sources are enlisted at the end of the article.
Related article:
- Replication 103: DNA Helicase- Structure, Function and Mechanism of DNA Unwinding.
- Replication 108: What is DNA Ligase and How It Helps in Replication?
Key Topics:
What is SSB Protein?
SSB stands for Single-Stranded Binding Proteins. As the name suggests their function is to remain attached to the single-stranded DNA. It is significantly found during the process of replication.
The eukaryotic SSB protein is also known as Replication Protein A (RPA). We can say the SSB and RPA are similar proteins.
It works downstream to DNA helicase and just before the replication fork. As the helicase unwinds the dsDNA, the SSB protein immediately binds with the single-stranded DNA.
The replication process is prevalent in all organisms, SSB is also present in prokaryotes, eukaryotes and viruses. Usually, a single SSB is a tetramer in structure with four different domains.
The structure of SSB is conserved across prokaryotes and eukaryotes with slight modifications. Note that SSBs are a class of nucleoproteins and are often known as DNA-binding proteins.
Structure of SSB:
Structurally, SSB proteins are assembled by various oligomeric subunits. For example, prokaryotes (E.coli) have four different subunits (tetramer). Eukaryotes have multi-subunit SSB proteins.
An SSB protein, in general, consists of OB fond (oligonucleotide binding fold), nucleic-acid binding domain, Cleft, and C-terminal tail, majorly. Other domains may also be present in various organisms.
A nucleic acid binding domain, as the name suggests, binds with the single-stranded DNA (nucleic acid). The positively charged amino acids present in this domain bind with the negatively charged single-stranded DNA.
Immediately after, the cleft (often known as DNA-binding cleft) ensures DNA-protein binding and tightens the interaction. Here comes the OB fold. The oligonucleotide binding fold is an important part of the SSB protein.
The fold contains 5-stranded beta sheets and an alpha-helix. The arrangement of OB-fold varies among bacteria, archaea, and eukaryotes. The SSB hallmark domain OB fold is responsible for validating DNA-protein binding and allows it for further processes.
SSB’s activity can be modulated by the C-terminal tail. This is the general overview of the SSB structure. Notedly, eukaryotes also consist of additional multifunctional domains for the replication protein A.
Related article: Comparison Between Helicase vs. Topoisomerase.
Types of SSB:
Viruses, archaea, bacteria, phage, plastids and eukaryotes, all organisms possess SSB proteins. And majorly, the function is to provide a stable and single-stranded DNA for replication.
Eukaryotic SSB protein also known as Replication Protein A (RPA) is a multi domain DNA binding protein. It not only works in DNA replication but also in DNA repair and recombination. RPA1, RPA2 and RPA3 are three important genes that encode various subunits of the RPA proteins.
The bacterial SSB protein, as aforesaid, is a tetrameric holo-structure. Entry of the ssDNA into a holo part presents DNA duplex formation. Again, the function is the same, to facilitate better replication, repair and recombination.
Viruses also contain a gene for their SSB protein. For example, the ICP8 protein of the HSV-1 (similar to the human cytomegalovirus) presents during the lytic cycle. What it does do is, destabilize the dsDNA, melt it and bind with it.
Phages also produce their own SSB protein upon host infection. Organelles like mitochondria or chloroplast also possess their own single-stranded binding proteins.
Mitochondria of yeast and eukaryotes encode its own SSB protein. In yeast, mtSSB protein is encoded by the RIM1 gene while human mtSSB protein is encoded by the SSBP1 gene.
In Saccharomyces cerevisiae, mitochondrial SSB protein functions in mitochondrial DNA replication. It is believed that RIM1 is exclusively responsible for ssDNA binding and can form homo-tetramers in solution.
Human mtSSB protein binds to single-stranded mtDNA as a tetramer. Thus, the human mitochondrial SSB protein has high sequence similarity with the E.coli SSB.
Related article: Organelle DNA- Mitochondrial and Chloroplast DNA.
Mechanism of Action:
The mechanism of SSB or RPA action is simple. Once the helicase unwinds the DNA, the SSBs recognize the single-stranded DNA. Various domains execute various functions during the process and effectively bind the SSBs to the single-stranded DNA.
Nucleic acid binding domains help in binding, DNA-protein binding cleft ensures correct binding and the OB domain validates the binding. Collectively, once the DNA is exposed to the SSB proteins, it remains in a single-stranded form.
Now, primase can use the strand to prepare the primer, while polymerase recognizes the single strand of DNA and starts DNA synthesis. SSBs have other functions as well.
Functions of SSB Protein:
Allow replication:
The pivotal function of either SSB or RPA is to allow replication. We already have discussed the mechanism of how it works during replication.
Protects from nucleases:
Single-stranded DNA is prone to nuclease attack. Nucleases are the class of enzymes that degrade the DNA. Once it finds the single-stranded DNA, it starts cleaving it. So here, in replication, it creates a problem.
Fortunately, SSP proteins bind with the DNA in multiple numbers, prepare a kind of ‘protective’ coat around the single-stranded DNA and prevent exposure to nucleases. Single-stranded DNA remains intact and replication can work efficiently.
Promotes DNA polymerase binding:
DNA polymerase is an enzyme that adds nucleotides during replication. However, it works only if it finds a single-stranded DNA. SSB proteins once bind with the ssDNA, polymerase recognizes the signal and prepares for synthesis. In addition, it also provides stability during the synthesis process.
DNA repair:
SSB proteins have a crucial role in the DNA repair process. The single-stranded DNA damage is prone to nuclease attack. SSB proteins bind with the single-stranded damaged region immediately and prevent nuclease action until helicase and DNA polymerase come to the rescue.
Interact with other proteins:
SSBs also interact with other proteins in various other processes like recombination and repair.
Other functions:
It prevents single-strand hardening and helps clear secondary structures from the single-stranded DNA, like loops or hairpins. This provides ease in replication.
Wrapping up:
SSB proteins or RPA are crucial elements of DNA replications. Broadly, it maintains our genome in a proper share and helps in inheriting genetic information from one to another generation.
In conclusion, An SSB protein is an important part of cellular machinery and helps in DNA replication, repair, recombination and maintenance. SSBs are now commercially available and widely used in various in vitro studies. We will discuss the applications of the SSB protein in our next article.
Sources:
Maffeo C, Aksimentiev A. Molecular mechanism of DNA association with single-stranded DNA binding protein. Nucleic Acids Res. 2017 Dec 1;45(21):12125-12139. doi: 10.1093/nar/gkx917. PMID: 29059392; PMCID: PMC5716091.
Guo JT, Malik F. Single-Stranded DNA Binding Proteins and Their Identification Using Machine Learning-Based Approaches. Biomolecules. 2022 Aug 26;12(9):1187. doi: 10.3390/biom12091187. PMID: 36139026; PMCID: PMC9496475.