“Genetic markers are known DNA sequences which can be used for studying any kind of polymorphism”.
In other words, we say,
A genetic marker is a known location on a chromosome used for the identification of individuals among and between species.
Thus from the definition, we can say that broadly, the Genetic markers are used,
- For studying disease gene
- For studying any kind of alterations (polymorphism)
- Identification of individuals
- Identification of species or organisms
In the present article, we will understand some common markers used in the molecular genetic technique along with their applications.
Content of the article:
- What is a genetic marker?
- RFLP(Restriction fragment length polymorphism)
- AFLP (Amplified fragment length polymorphism)
- RAPD(Rapid amplified polymorphic DNA)
- STR (Short tandem repeats
- ISSR(Inter simple sequence repeat)
- SCAR (sequence characterized amplified region)
- EST (Expressed sequence tag)
- SNP(single nucleotide polymorphism
- SSR(simple sequence repeat)
- Applications of genetic markers
- Conclusion
Key Topics:
What is a genetic marker?
“The genetic markers are often known as DNA marker is a known DNA sequence located in the genome, used in the molecular genetics for identification of alteration or DNA sequence from the unknown DNA sample.”
Or
The genetic marker is a known DNA sequence or gene located on the chromosome which can be applied in the identification of individual species or organisms or we can use it in the identification of other genes or DNA sequences.
Before that let me give you a brief idea about how different genetic markers are developed.
If we mark some sentences or paragraphs, we can use them every time in exams, assignments and other activities.
Similarly, by marking a specific sequence of DNA, we can use it in different types of genetic studies.
Let’s take an example:
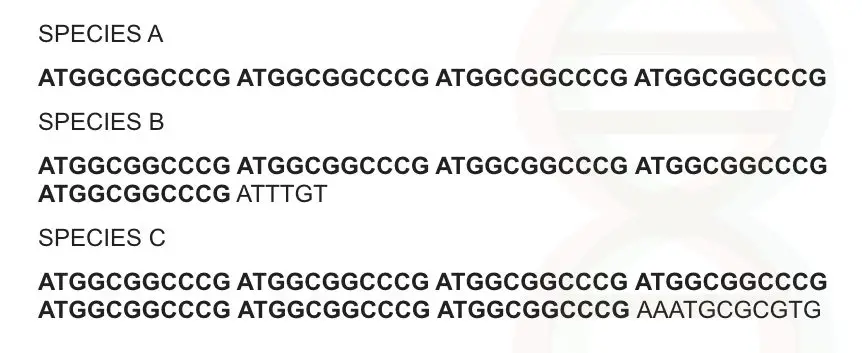
Can you identify something? 11 basepair sequence is repeatedly observed at several intervals in a sequence and it is present in all three species.
Suppose this sequence is abundantly present in all organisms on earth (assume it). Let me give some name to this marker by using its characteristics.
It is observed one after another, tandemly.
It is repeatedly observed after a regular interval of sequences hence it is a repeat sequence.
Though the sequence is the same, sequence numbers are different in all three species i.e, 4 repeats in species A, 5 repeats in species B and 7 repeats in species C. So the number of repeats is variable.
Now collect all three characters in a single set: it is a sequence, arranged tandemly, repeated one after another and variable so we can name it as variable numbers of tandem repeats.
That is how different markers are developed.
RFLP: Restriction fragment length polymorphism
With the help of the Restriction digestion by REase, alterations in the same/ homologous DNA sequences can be detected by analyzing fragments of different lengths, digested with a restriction enzyme.
A single restriction endonuclease gives more specific results by cutting at one specific locus and producing fragments of different lengths. See the figure on how different DNA fragments are created by Restriction digestion.
Figure,
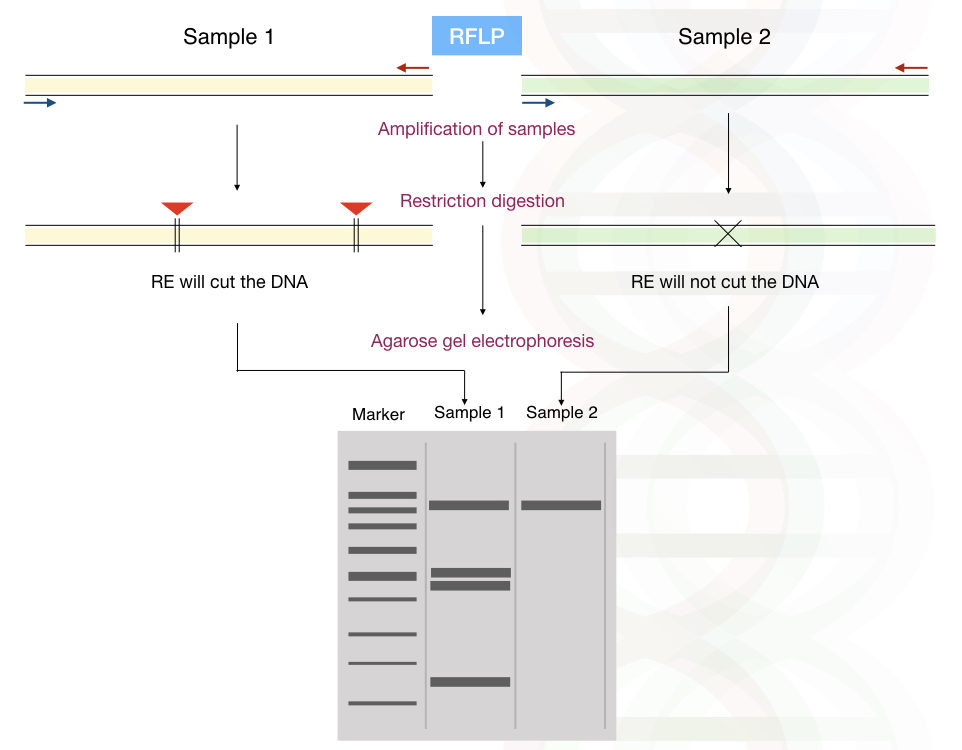
Firstly, let us understand the terminology RFLP, alteration (polymorphism) in the length of different fragments (of DNA) can be analyzed using restriction digestion.
Restriction digestion is performed by endonuclease enzymes. It cuts DNA at its specific restriction site. Millions of restriction sites are present for individual RE in the human genome.
The length of different fragments is identified using blotting, which is now replaced by sequencing. RFLP is applicable in disease identification, genetic mapping, heterozygous detection and carrier identification.
The RFLP markers are highly locus-specific and co-dominant.
Why RFLP markers are locus-specific?
Because of the nature of the REase used in the rflp.
As we discussed in the previous article, the REases can cut at a specific location where their restriction site is located. One restriction digestion site generates two different fragments.
Further, it is co-dominant because both alleles in the heterozygous condition (mutant as well as normal) can be detected by rflp.
There are two variations in the RFLP that are most common.
The traditional method of the RFLP is the blotting-based probe hybridization method.
The PCR-based RFLP method is a newer version, easy and rapid.
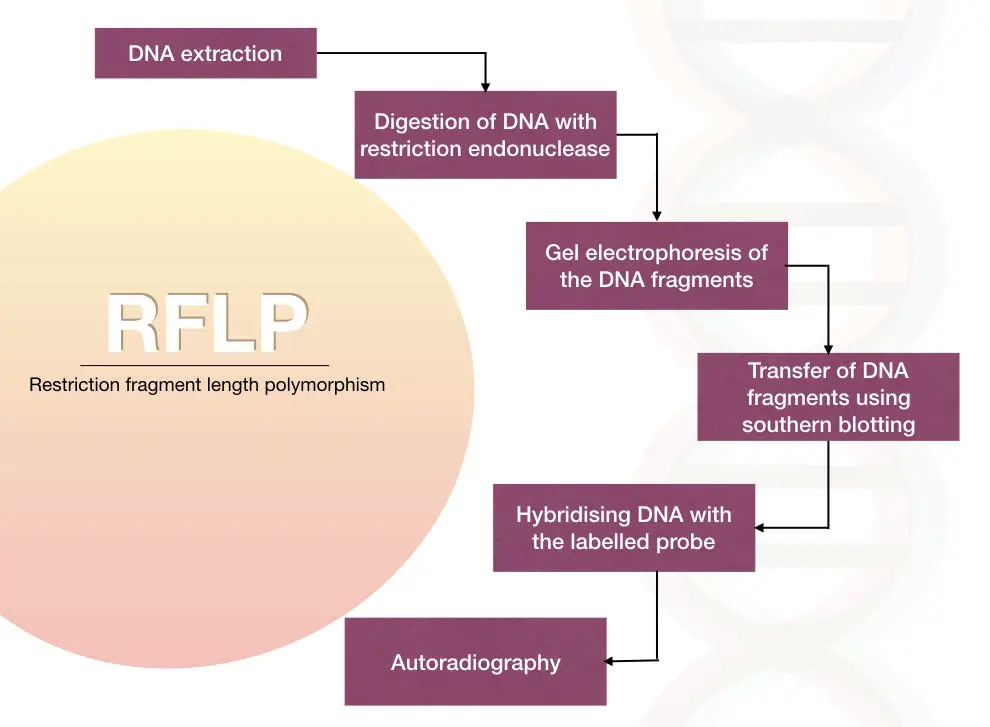
In the traditional method, the labeled DNA probes called an RFLP probe, used to hybridize with the digested fragments by the Southern blotting analysis.
The polymorphism can be detected by analyzing the hybridization pattern of the labeled probe using autoradiography
In the PCR-based RFLP method, the gene of our interest is amplified using the polymerase chain reaction and proceed for the restriction digestion using the specific REase.
The results are analyzed using agarose gel electrophoresis.
No tedious labeling and hybridization techniques are required in this method.
The PCR-RFLP method is easy, simple, time-efficient and reliable.
Polymorphism in the alleles of a gene such as MTHFR and HBS can be detected using PCR- RFLP.
Application of RFLP:
It is used in polymorphism study, forensics, paternity verification, hereditary disease diagnosis and DNA fingerprinting and genome mapping.
AFLP: Amplified fragment length polymorphism
The AFLP technique was developed by Zabeau and Vos in the year 1993. The method is based on the amplification of the selected fragments followed by the digestion of the total genomic DNA of the selected organism.
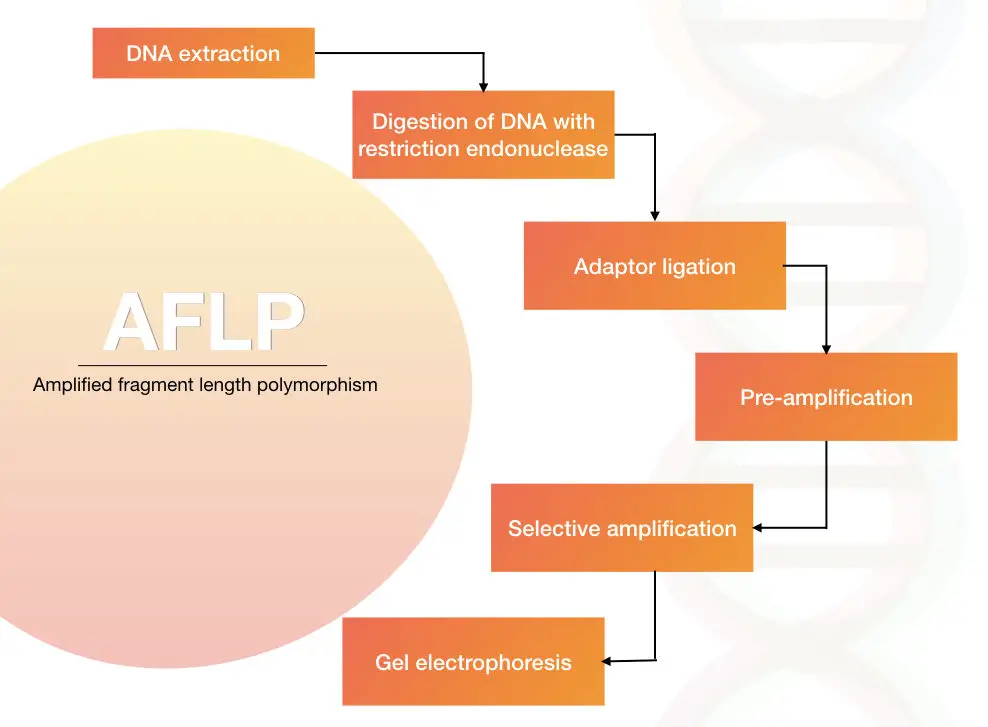
The process of the AFLP is restriction digestion of the genomic DNA, adapter ligation of the digested DNA fragments and amplification of DNA fragments in PCR using the adapter specific primers.
The method was originally developed by the keygen in 1990.
The genomic DNA is extracted using one of the DNA extraction methods.
The DNA is digested with the help of the restriction endonuclease
The digested DNA is ligated with the short adapter sequences. The sequence of the adapters is known.
The fragments are amplified using a set of primers.
The results are analyzed on agarose gel electrophoresis followed by autoradiography or we can use the PAGE technique to differentiate multiple fragments of DNA.
The AFLP technique is highly applicable in Crop improvement programs and genomic analysis of different types of crop varieties.
As the primers are based on the adapter data, no previous data on the DNA sequences are required to construct a marker.
Due to the high priming capacity of the primers, it is also used in phylogenetic studies for different plant varieties.
Advantages:
- No prior sequence information is required in the AFLP.
- Also, the sensitivity, reproducibility and resolution of the AFLP marker are higher as compared with other markers.
Disadvantages:
- The AFLP marker is not co-dominant hence it can not distinguish homozygous from heterozygous.
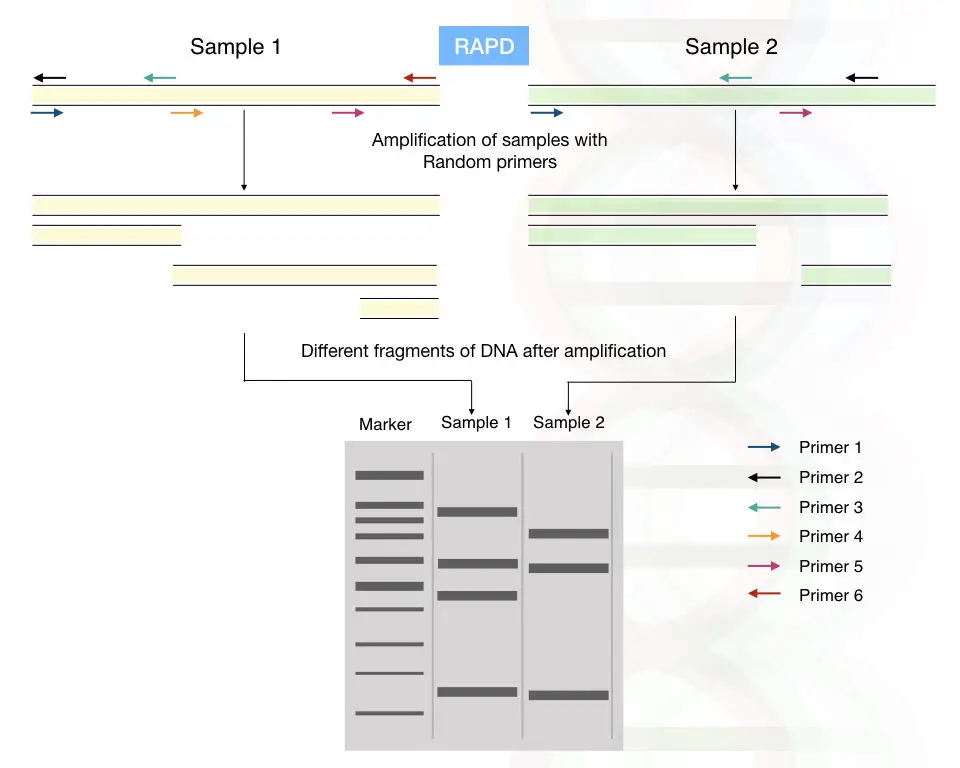
RAPD: Random amplification of polymorphic DNA
By using the short, arbitrary DNA primers, the part of the genomic DNAs is randomly amplified.
Pronounced as a “rapid”, it does not require any previous sequence information, the primer of 8 to 12mer randomly amplifies the region of the genomic DNA.
If any mutation or alteration occurred in the region of the primer binding site, the primer can not bind to that location or the amplified fragment length decreases or increases.
A different pattern of amplification is observed in different sample types. Hence the RAPD can help in the polymorphism studies.
Detection of RAPD:
The RAPD fragments are between 0.2 to 5.0kb. It can be observed using the ethidium bromide-stained agarose gel or it can also be analyzed on polyacrylamide gel electrophoresis.
Advantages:
- The RAPD markers are abundantly present in the genome and are distributed throughout the genome.
- No previous sequence information is needed.
- It can amplify a low quantity of DNA.
Disadvantages:
- The marker is not locus-specific.
- The sensitivity of the RAPD is also lower.
- The reproducibility is also very low.
- Homozygous and heterozygous are not easily distinguished.
- The RAPD results are difficult to interpret because the short primer can amplify any of the random sequences presented into the genome.
Nonetheless, RAPD markers are quite popular in gene mapping, species identification, species-specific polymorphism studies and phylogenetic analysis.
It is popular in plant genetic research.
ISSR: Inter simple sequence repeat
The ISSR marker is the inter simple sequence repeat, the method is based on the PCR agarose gel electrophoresis, detected using the single primer amplification reaction.
Between the oppositely oriented microsatellites, the sequence of 100 to 3000bp fragments of a sequence is called the ISSR.
Here the advantage of using the ISSR is that no prior sequence information is required for constructing the primers.
Multiple loci are amplified because of the random distribution of ISSR within the genome.
Also, the low quantity of templates is sufficient to do PCR.
However, it has lower reproducibility and non-homology of similar-sized fragments.
Because of the simple setup of the ISSR, it is employed in gene mapping studies, clone identification, different strain identification and parental verification.
It is a highly polymorphic marker and is used in genetic diversity studies, gene tagging, evolutionary study and phylogenetic studies.
In this technique, the flanking non-repeated regions of the inversely oriented microsatellites are amplified using a single primer or pair of primers.
STR: Short tandem repeats
The short tandem repeats/ simple sequence repeats are also called a microsatellite. The microsatellite is the shorter repeats sequences present into the genome thousands or millions of times.
In other words, we can say that the STRs are tandem repeats but are much shorter than the VNTR. The repeats in STR are 2 to 6 base pairs per repeat.
The STR is highly polymorphic, the number of repeats differs between individuals even though the loci are homologous. Because of this reason, the STR marker is the first choice in forensic analysis.
Due to the difference in the repeat number each individual has a different STR profile.

Read more on STR: Short Tandem Repeats (STRs): A Secret of Every DNA Test.
SCAR: Sequence characterized amplified region
The SCAR stands for sequence characterized amplified region.
It is easy to use, reliable and reproducible.
The present marker is based on the PCR-agarose gel electrophoresis, uses longer primers of 15-30 nucleotides hence the reproducibility increases anonymously.
It is a locus-specific and co-dominant marker.
Unlike the RAPD, it required sequence information for designing the primer for SCAR.
It is widely used in plant marker assistance selection studies.
EST: Expressed sequence tag
Expressed sequence tag is a 500 to 800bp sub-sequence of a short stretch nucleotides of the expressed gene or the cDNA.
A small segment of the entire cDNA library is sequenced for the generation of the EST.
The EST marker is actually a short stretch of the individual cDNA, which represents a tag for the entire cDNA.
This tag can be used in the study and identification of the entire gene, thus is no need to sequence an entire gene.
We will discuss more on the EST in some other articles.
SNP: Single nucleotide polymorphism
SNP is a single nucleotide polymorphism, a type of unique marker in comparison with other markers enlisted here.
Due to the addition or deletion of a single nucleotide in a genome, the SNP originated.
Millions of SNPs are present in the genome, however, they may or may not be pathogenic.
The SNP marker is used in the Genome-wide association studies.
By comparing the case and control groups, the association of a particular SNP associated with a particular disease can be determined.
For more detail read our article on SNP: An Introduction To Single Nucleotide Polymorphism (SNP).
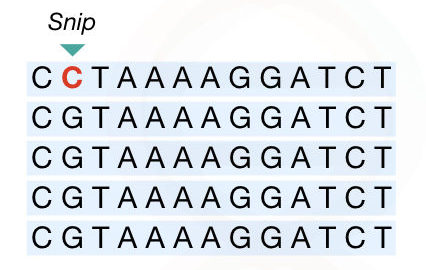
SSR: Simple sequence repeat
A simple sequence repeat is a microsatellite repetitive DNA marker, typically ranging from 5 to 50 times in a genome.
The marker is highly variable. It is more similar to the short tandem repeats. We had discussed STR in the above segment.
The SSR marker or the microsatellite marker is co-dominant and restriction digestion is not involved in this process thus, advantageous over the RFLP.
It is a marker based on the polymerase chain reaction.
Applications of genetic markers:
The genetic markers are employed in the identification of traits related to disease or inherited abnormal conditions.
The genetic markers are also utilized in the identification of species, individuals or organisms.
Thus the DNA markers like the VNTR and STRs are one of the first choices in DNA fingerprinting, parental verification and maternal cell contamination.
It is also applicable in population studies and genetic variations studies.
The markers like RAPD and SSCP have great value in plant genetic studies such as plant hybridizations studies, DNA barcoding, plant breeding, creating stress-resistant species, etc.
Furthermore, genetic markers can also be used in medicinal plant research and phylogenetic analysis.
It is frequently used in marker-assisted breeding programs.
RFLP, SNP and AFLP like genetic markers are now routinely used in genetic disease analysis and diagnosis.
Furthermore, gene mapping and genetic screening can be done using genetic markers.
The techniques like AFLP have great value in the characterization and identification of genetic variations in closely related species.
Besides these several other applications are:
Quantitative analysis of bio-sample.
Species and speciation studies.
Sex determination can be done using the Y-specific markers.
Quantitative trait loci identification and characterization.
Seed testing.
Marker-assisted selection.
Cloning and gene transfer experiments.
The SNP-like genetic markers have great value in genome-wide association studies.
Conclusion:
The genetic markers are one of the best choices for location disease-causing genes on a chromosome. Furthermore, cancer genetic markers are nowadays used in the diagnosis and prognosis of cancer.
Read our article on Cancer genetics: A Brief Introduction To Cancer Genetics.
Sources:
- Young, N.D. Plant Growth Regul (1993) 12: 229. https://doi.org/10.1007/BF00027203
- Raza. S., Shoib. W. and Mubeen H. “Genetic markers: importance, uses and applications.” International Journal of Scientific and Research Publications; 2016: 6(3): 221-223. Link.


