“The 16S rRNA gene is a bacterial ribosomal gene and a part of the 30S subunit which is used in the identification, characterization, and classification of various bacteria.”
16S rRNA gene constructs the 16S rRNA subunit which binds to the Shine-Dalgarno sequence present in the bacteria genome. Although the gene is highly conserved since evolution and therefore it is often known as a “molecular fossil”.
Notably, the gene has two different domains, a larger one conserved domain, and a hypervariable region. Each bacteria has different hypervariable sequences.
In the whole solar system our earth has a life with two most common forms; prokaryotes and eukaryotes. The prokaryotes are single-cell organisms such as viruses or bacteria while the eukaryotes are multicellular and have different degrees of cellular organization.
It was believed that every present life form was evolved from the primitive prokaryotic organisms. Bacteria are the most common prokaryotic life form, some are harmful and some are useful.
For instance, lactobacillus and some gut bacteria are helpful while some strains of E.coli and C. botulinum are harmful to us.
Using microbiological techniques various kinds of bacteria can be identified. However, conventional culturing and microbiology methods are time-consuming, prone to contamination, and less accurate. It takes a few days to culture bacterial cells.
Also, some bacterial strains are difficult to culture and some are highly infectious and lethal to us.
Thanks to the recent genetic advancements, using the state of the art molecular techniques, bacterial identification, classification, and other characteristics can be studied well.
Methods like different forms of PCR and DNA sequencing are accurate, efficient, fast, cost-effective, and contamination-free compared to the conventional microbiological methods. Moreover, within a few hours, we can get results.
The present article is more valuable for a microbiology student than others. However, this article will definitely help every biotechnology student. In the present article, I will try to let you understand what the 16S rRNA gene is and how it’s sequencing is done.
Related article: Microbial genetics.
Key Topics:
Introduction to 16S rRNA:
In 1977, Carl Woese and George E Fox had used the 16S rRNA gene in bacterial phylogenetic analysis. This was the first attempt to do so. Their findings and use of genetic techniques had revolutionized the field of microbiology.
As I explained, it is a subunit of a 30S ribosome RNA, It plays a remarkable role in bacterial translation. Without it, the ribosomal assembly can’t function correctly and the ribosome can’t function efficiently.
Let’s see some of the key functions of the 16S rRNA in a biological system:
Function:
It interacts with the other ribosomal subunits like 23S and facilitates bindings of 50Sand 30S ribosomal subunits.
As it is a major structural support to the ribosome, it helps in establishing ribosome functioning. It also gives structural support to the ribosomal protein by operating as a scaffold.
The 3’ end of it also helps in the initiation of protein synthesis by interacting with S1 and S21 protein.
At the 3’ end, the 16S rRNA has some special kind of sequences known as anti-Shine-Dalgarno sequences that have the capacity to bind at AUG of mRNA.
At the A-site of the ribosome, It stabilizes the correct codon-anticodon pairing.
Conclusively, the present RNA performs a very vital role in synthesizing prokaryotic proteins.
16S rRNA gene:
It is evident that some genes form 16S rRNA. But immediately one question arises in your mind that, it is a type RNA and genes form proteins, how is it possible?
As per the traditional definition of a gene, “a functional unit of a genome that forms protein is known as a gene.”
But, as per recent findings, genes form not only proteins but also construct various RNAs. These types of RNAs are rRNA, tRNA, miRNA, siRNA, shRNA, and other non-functional small RNAs.
Notably, these RNAs can’t be translated into protein, unlike the mRNA but it plays an essential role in transcription and translation regulation. Thus these RNAs are further involved in gene regulation.
The 16S rRNA gene has two important domains, one is the conserved domain, present in all bacterial species, and is unchanged. While the other domain is a hypervariable region. See the image below,

The hypervariable regions provide exceptional diversity to bacteria. It is thus variable, different bacteria have different alterations of it which makes each bacterium unique. 9 different hypervariable regions V1 to V9 are usually present in a gene.
As the hypervariable region is a kind of blueprint for every bacteria, it is widely used in identification, classification, detection, comparison, and phylogenetic analysis of various bacteria.
5 to 10 copies of the 16S rRNA gene are present in a single bacterial cell which makes the detection sensitivity higher. The gene has around 50 various functional domains with a total length of up to 1550bp.
For detection, the universal primer set is constructed based on the information of conserved domains (because it is present in all bacteria and is unchanged since evolution) while to detect specific species of bacterial or a bacterium, probe or another set of primer is designed based on the sequence information of the variable regions.
[epcl_box type=”notice”]The 16S rRNA gene is a kind of “molecular clock” used for phylogenetic analysis of various bacterial species and sea archaea.[/epcl_box]
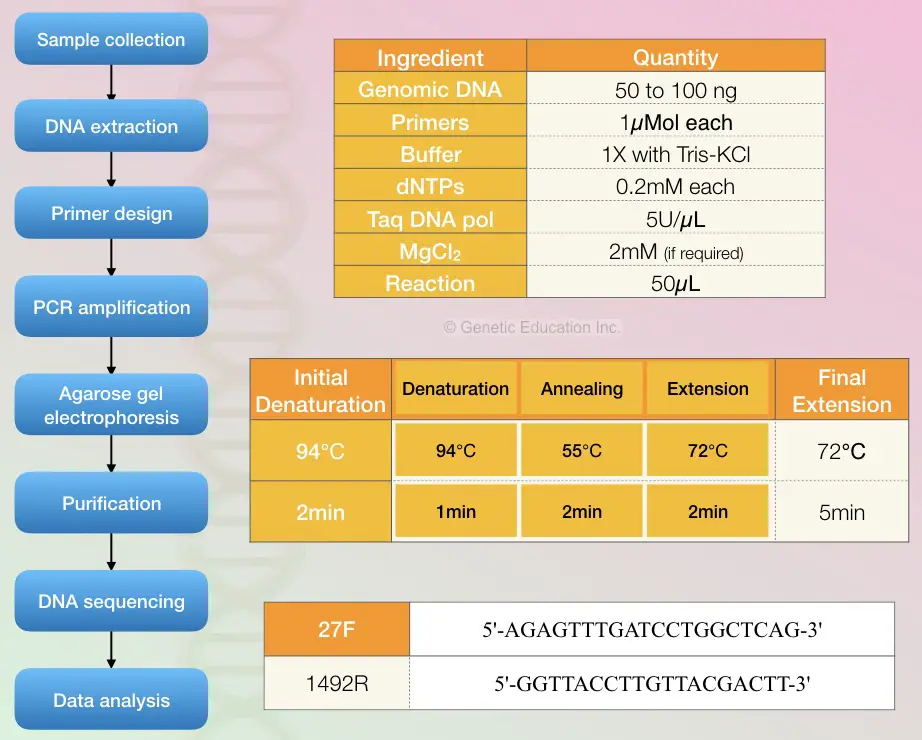
Different sets of primers are now available to do PCR using 16S rRNA however, the primer set 27F and 1492R used by Weisburg et al are the most common primer set used in the study.
Related article: What Is A Gene?- Definition, Structure and Function
Nonetheless, to reduce the reaction complexity we can design our own set of primers to make shorter PCR amplicons. Some of the common primer sets used for PCR based detection are enlisted in the table below,
| Primer name | Sequence (5′–3′) |
| 8F | AGA GTT TGA TCC TGG CTC AG |
| 27F | AGA GTT TGA TCM TGG CTC AG |
| 928F | TAA AAC TYA AAK GAA TTG ACG GG |
| 336R | ACT GCT GCS YCC CGT AGG AGT CT |
| 1100R | GGG TTG CGC TCG TTG |
| 907R | CCG TCA ATT CCT TTR AGT TT |
| 785F | GGA TTA GAT ACC CTG GTA |
| 805R | GAC TAC CAG GGT ATC TAA TC |
| 533F | GTG CCA GCM GCC GCG GTA A |
| 518R | GTA TTA CCG CGG CTG CTG G |
| 1492R | CGG TTA CCT TGT TAC GAC TT |
Detection methods:
PCR and sequencing are two common methods scientists are using to detect bacterial species. Although the PCR (polymerase chain reaction) has some detection limitations, we will talk about it at the end of this section.
PCR:
The PCR is a temperature-dependent amplification method that synthesizes DNA sequences. Denaturation of template, annealing of primers, and extension of a sequence occurs in three different steps, viz denaturation, annealing, and extension, respectively.
Here, a set of primers specific to the conserved domain of a bacterial gene with other PCR ingredients like PCR buffer, dNTPs, Taq DNA polymerase and nuclease-free water are used to amplify target gene.
Once the amplification is completed, the reaction is run of the agarose gel to detect the amplification. The presence of amplicons or DNA bands indicates the presence of bacterial strain. While no amplicons indicate the absence of bacteria.
Broadly, it gives us an idea about the presence and absence of bacteria in any sample. However, novel bacterial strains or species can’t be detected using PCR.
If we have sequence information or a target to study, we can do so, otherwise, we have to do DNA sequencing.
In addition to this, PCR can be used in quantification of the bacterial load as well. The amount of infection, bacterial load, or quantity of bacteria can be determined using the real-time PCR method.
The 16S rRNA gene is also used to know the relatedness of organisms and evolutionary distance between organisms.
DNA sequencing:
DNA sequencing is a method used to identify the sequence of DNA present in any biological sample. Here we are using labeled nucleotides or probes to know the DNA sequence.
Notably, instead of whole bacterial genomic DNA, amplified 16S rRNA gene is utilized as a template for sequencing. As the labeled dNTPs bind to the sequence, a detector detects the fluorescence emitted by it.
Once the entire sequence is completed, using comparative analysis, the 16S rRNA gene sequence is compared to other sequences to distinguish any variations if any. Doing so, we can create a phylogenetic tree of various bacteria.
We can sequence the entire 1500bp region of a gene but interestingly, the first 500bp region is potent enough to establish differences between various bacterial species or taxa. Appx. 66% of 16S rRNA gene variations are in the first 500bp (studied in Bordetella species).
Read more on DNA sequencing: DNA Sequencing: History, Steps, Methods, Applications and Limitations.
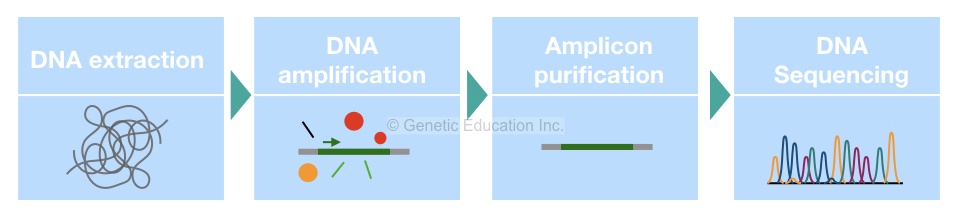
16S rRNA detection: Complete process
The first step in the detection process is sample collection and DNA extraction. Routine DNA extraction protocols like PCI or proteinase K can’t work here, we need to use a standard bacterial DNA extraction protocol or the ready to use DNA extraction kit to obtain the bacterial DNA.
In the next step, quantify the DNA and check the purity of extracted DNA. Select only the good quality DNA, if not obtained, repeat DNA extraction.
In the next step, design or select the primer pair for PCR amplification. Usually, a single set of primers specific to the 16S rRNA gene is enough to do amplification. Select only the 500bp amplification region. However, as the gene is too small we can also amplify the entire gene as well.
The reaction preparation process and quantity of each ingredient are given into the figure below. Also, the cyclic PCR conditions are given in the figure below.
PCR protocol for 50microliter PCR reaction.
- Genomic DNA: 50 to 100ng
- Primers: 1micromol
- Reaction buffer 1X (Tris-KCl)
- dNTP mix: 0.2mM
- Taq DNA polymerase: 5U/microliter
- MgCl2: 2mM (if required)
PCR cycling conditions:
- Initial denaturation: 94C for 2 minutes
- Denaturation: 94C for 1 minute
- Annealing: 55C for 2 minutes
- Elongation: 72C for 2 minutes
- Final extension: 72C for 5 minutes
- Perform PCR for 30 cycles.
Once the reaction is completed, confirm the complications on the agarose gel electrophoresis. Use only 2% or 1.8% agarose to do so. The entire process of agarose gel electrophoresis is given into the article below,
Now in the next step, purify the amplicons again using the ready to use DNA purification kit and send it for DNA sequencing.
Using a set of primers and probe the sequencing machine identifies the whole sequence of a 16S rRNA gene. After completion of the sequencing process, the data are sent to the bioinformatics lab for the evaluation of the results.
Now in the computational analysis, the entire sequence of the 16S rRNA gene is compared with the available sequence. An expert identifies alterations or variations in the hypervariable region of a gene and tries to find out its relation with other available sequences.
[epcl_box type=”information”]GeneBank has 90,000 different 16S rRNA gene sequences of various bacterial strains. [/epcl_box]
Applications of 16S rRNA:
- Identification of bacteria species
- Taxonomic analysis
- Characterization of a bacterial population
- Assessing bacterial diversity
Conclusion:
PCR and DNA sequencing help scientists to do microbial identification accurately and fast. Nowadays various ready to use kits based on PCR amplification are available to boost the automation process. However, using the manual method can increase knowledge and decrease the cost of reaction.
Sources:
- Ota-Tsuzuki C, Brunheira AT, Mayer MP. 16S rRNA region-based PCR protocol for identification and subtyping of Parvimonas Micra. Braz J Microbiol. 2008;39(4):605‐607.
- Widmer F, Seidler RJ, Gillevet PM, Watrud LS, Di Giovanni GD. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol. 1998;64(7):2545‐2553.