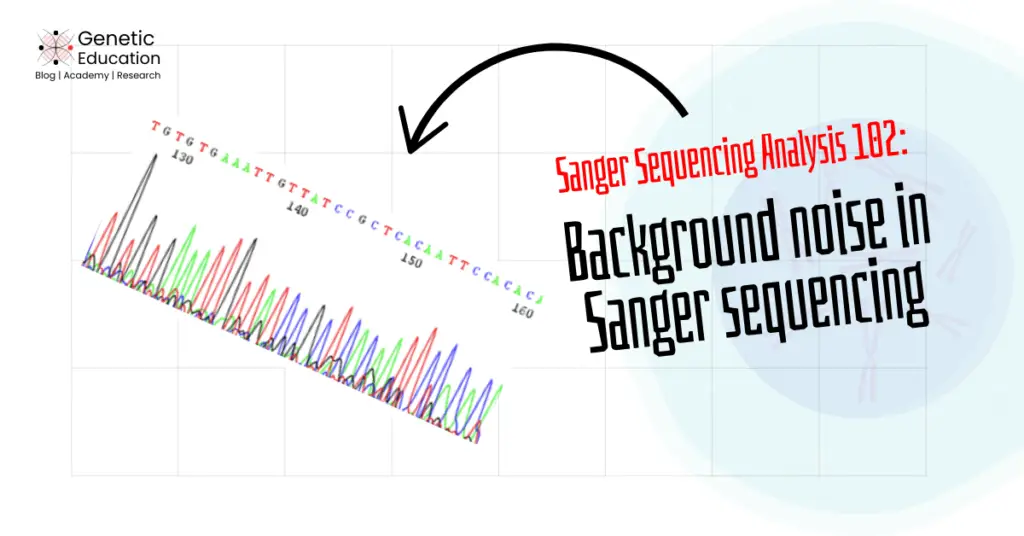
“Background noise is smaller undefined peaks beneath our main sanger sequencing peaks. It can be generated for various reasons and can also be resolved. Learn about background noise and troubleshooting in this article.”
Sanger sequencing is the gold-standard technique for studying the DNA sequence so far. The advances as automation in Sanger sequencing, lead us to keen observation and data analysis.
Sequencing peak generation by the fluorescence emission helps investigate the sequence even more accurately. However, there is one problem in this— background sequencing noise.
Imagine, you are trying to study a photograph or painting, but there are so many unwanted elements cluttering the image that it ruins your focus. That’s exactly what background noise does in Sanger sequencing—it disrupts clear observation, making it difficult to analyze the DNA sequence accurately.
Overall, it makes the results least trustworthy and undeliverable.
This is the second article of our sequencing analysis series and in this article, I will explain the background noise, how it is generated and how to overcome it.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Key Topics:
What is Background noise in sequencing?
The trace (rough) file of Sanger sequencing shows peaks for each nucleotide. Any unwanted peaks or signals with these peaks are considered as “Noise” here, Not the actual noise!
Usually, the trace file should show clear, distinct and sharp nucleotide peaks, however, unwanted signals make the background of the sequencing file ‘noisy.’ And thus called background noise.
Any unwanted, smaller, low-quality signals within the sequencing trace file are known as background noise.
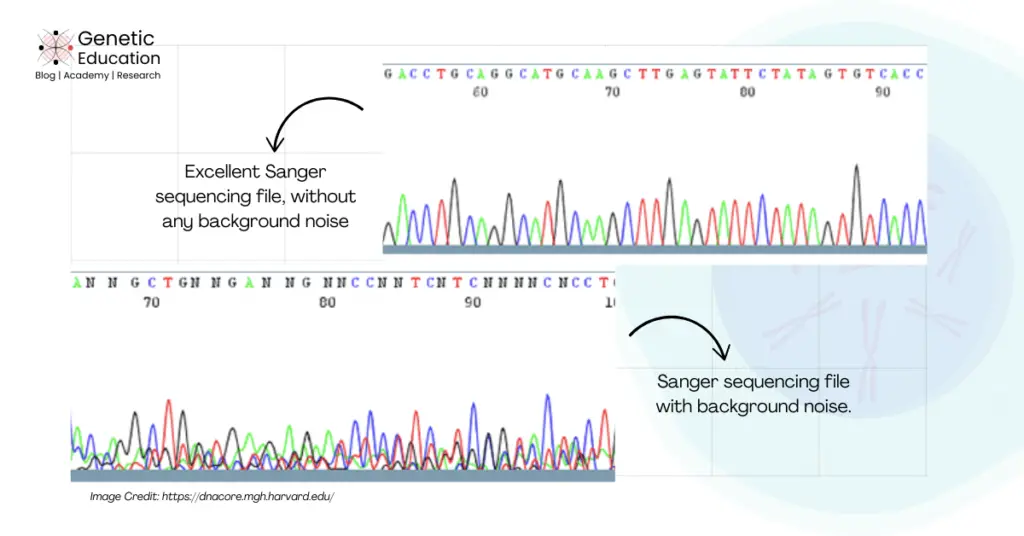
Keep in mind that too strong background noise creates difficulties for the software to resolve the nucleotide. It will mislead the automated base caller or software and call the wrong nucleotides throughout the sequencing trace.
Technically, background noise is smaller, undefined and unidentifiable peaks or signal traces present in the sequencing trace file. These signals are
- Smaller in size,
- Appear within the electropherogram, between or underneath the peaks,
- Appear at the beginning and end part of the sequence file,
- Appear only at certain parts or entire sequencing file,
- Appear undefined and ununiformly.
Types of background noise:
Various types of background noise are explained here.
Baseline noise:
Smaller, low-intensity peaks that appear in the chromatogram are known as baseline noise. Baseline noise is a type of sequencing artifact. It can be avoided during the analysis and usually doesn’t majorly impact the analysis part.
Weak signals:
Low and unclear peaks are considered weak signals and are also categorized into background noise. Weak signals appear due to poor-quality templates, poor dye quality or resolution, low DNA concentration and technical issues.
Weak signals significantly influence the electropherogram analysis part.
Dye artifacts:
As we know, four different fluorescent dyes have been used to label four different ddNTPs. Fluorescence signals are used to generate peaks. Sometimes, the dye can degrade or over-fluorescence with other dyes.
This generates unwanted smaller peaks in the trace file and is considered as background noise. Dye artifacts are also the most common cause of background noise in Sanger sequencing.
Noise at the end:
Smaller fragments are difficult to resolve, so at the end of the sequencing file, significantly high background noise will be observed. This, however, can not be overcome as it is a common sequencing artifact.
In the practical procedure, the end part of the sequence (up to 50 nucleotides) isn’t considered in the analysis and is thus removed.
N + 1 or N – 1 peaks:
N+1 and N-1 peaks, appearing unwantedly before and after the actual nucleotide peak, are also categorized into the background noise. Keep in mind, this type of background noise makes the whole analysis part difficult.
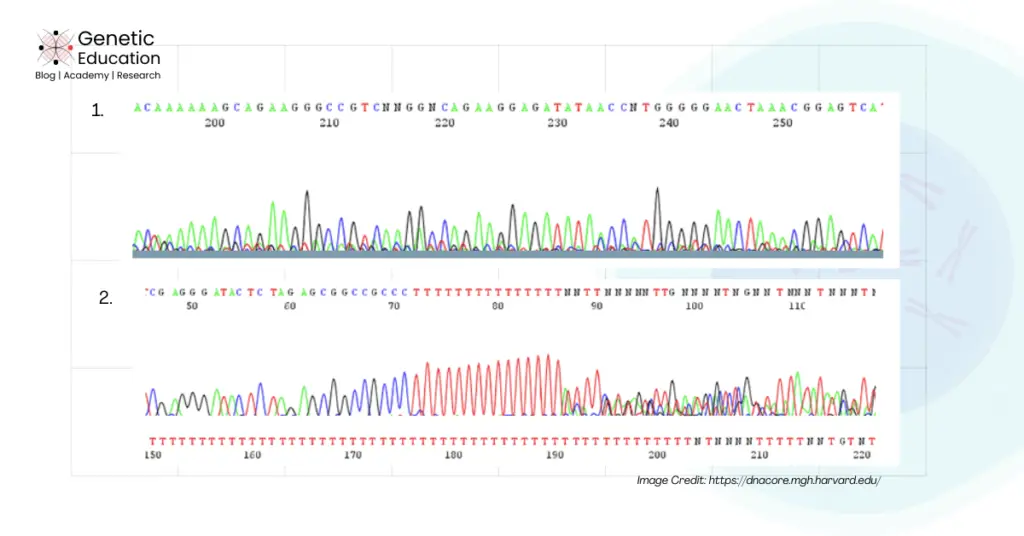
How is background noise generated?
Due to multiple reasons, various types of (discussed above) background noises are generated. Some of the common reasons are explained here.
Poor quality DNA:
Poor-quality DNA is the main reason for the generation of background noise. DNA with impurities, other DNA contaminants, containing amplification inhibitors, and degraded DNA can be considered as poor quality DNA.
Now what will it do?
It makes the amplification difficult, generates non-specific amplification, hurdles the target DNA amplification, or inhibits the amplification. Resultantly, it generates low-quality and secondary peaks in the trace file.
Related article: Template Preparation for Sanger Sequencing.
| Type of poor-quality DNA | Impact on PCR or sequencing | Background noise generated |
| Degraded DNA | Results in incomplete or no amplification, leading to low yields and poor sequencing data. | Produces weak or absent signal peaks, and irregular peaks in chromatograms. |
| Contaminated DNA | Inhibits polymerase activity, leading to inconsistent amplification. | Causes mixed or noisy signals, and multiple peaks in sequencing data. |
| Inhibited DNA | PCR fails to amplify effectively, resulting in low or no product. | Generates weak or no signals, poor amplification artifacts. |
| Low concentration DNA | Insufficient template for amplification, causing poor sequence coverage. | Causes random background noise, weak signals, or failed reactions. |
| Cross-contaminated DNA | Leads to the amplification of non-target sequences, reducing specificity. | Creates false-positive peaks, overlapping peaks in chromatograms |
| Sheared DNA | Results in fragmented amplification products, compromising sequence integrity. | Causes shortened reads, broad or uneven peaks. |
| High-GC content | Difficulty in amplification due to secondary structures or polymerase stalling. | Leads to drop-offs, low signals, or unresolved regions in sequencing. |
| Formalin-fixed DNA | Causes extensive crosslinking and fragmentation, leading to poor amplification. | Generates noisy data, fragmented peaks, or missing data regions. |
Amplification errors:
Amplifications have been a well-documented cause for the background noise generation in the sequencing. Common amplification or PCR errors like non-specific amplification, primer-dimer or multiple ex-template amplification result in moderate to high background noise.
| Amplification error | Explanation | Background noise |
| Primer-dimer | Primers bind to each other instead of the target sequence. | Short, unwanted fragments; extra peaks in chromatograms |
| Non-specific amplification | Amplification of non-target region. | Short, unwanted and uneven peak throughout the electropherogram. |
| Contaminant amplification | Non-target DNA gets amplified, usually due to contamination. | Mixed or overlapping peaks; unintended sequences. |
| Low DNA | Random amplification variation due to low DNA template concentration. | Uneven peak intensities; random noise. |
| PCR contaminant | Leftover dNTPs, buffer, polymerase and other ingredients also influence sequencing. | This can generate moderate to high level of background noise. |
Notably, non-specific amplification, primer-dimer and PCR contaminants are the common cause of background noise during Sanger sequencing. It is advisable to validate PCR amplicons and purify them before sanding it for sequencing.
Dye artifacts:
Dye artifacts are yet another notable reason that produces a significant level of background noise during Sanger sequencing. Dye incorporation error, overlapping, incomplete termination and residual dye signaling are the common dye artifacts reported.
| Dye artifact | Explanation | Background noise |
| Incorporation errors | Excess or improperly incorporated fluorescent dyes lead to non-specific signals. | Misleading peaks in the chromatogram. |
| Overlapping signals | Multiple dye labels with overlapping emission spectra cause interference in signals. | Composite peaks complicate data interpretation. |
| Residual dye | Unremoved unincorporated dyes after cleanup contribute to unwanted signals. | Mimics true sequence data, obscuring actual sequences. |
| Signal saturation | Excessive dye incorporation saturates the detection system, leading to flattened peaks. | Obscures genuine sequence information, resulting in inaccuracies. |
| Dye degradation | Prolonged exposure to light degrades fluorescent dyes, causing inconsistent signal intensity. | Spikes or drops in sequencing data. |
Incomplete termination:
N+1 and N-1 peaks nearer to the main nucleotide peak, are generated mostly due to incomplete termination.
Now, here what happens;
ddNTPs terminate the sequencing chain. Sometimes, the reaction doesn’t terminate at a correct nucleotide or position leading to partial unwanted extension. This generates additional peaks and results in background noise.
Polymerase errors:
Polymerase also produces errors during the synthesis process and contributes to significantly high background noise. Polymerase slippage in the repetitive regions and addition of wrong nucleotides are the major causes.
Technically, polymerase errors generate insertion or deletion type additional peak, and are actually background noise.
Signal degradation:
Signal degradation is almost always reported in every electropherogram at the beginning and end of the sequence. Henceforth, substantially high background noise is reported at the beginning and end of the sequence.
There are two reasons for these two artifacts.
Smaller fragments are difficult to resolve as they migrate faster. While larger fragments are resolved at the end with the least quality. In both cases, background noise is reported.
It is recommended to trim up to 50 nucleotides at the beginning and end of the trace to avoid these unresolved nucleotides.
Technical errors:
Lastly, a few types of technical and instrument errors can also produce continuous background noise. These are poor calibration, optical interference, electrical or instrumental problems etc.
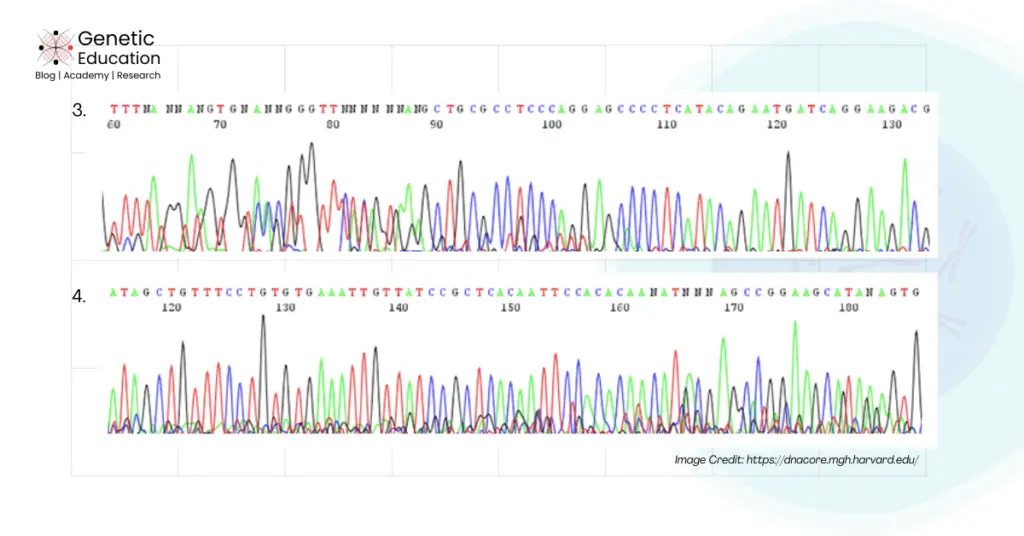
How to Reduce background noise in Sanger sequencing?
Now in this section, I will give you stepwise troubleshooting for background noise and lastly, I will also give you some of the common tips that will mostly work for you!
Poor quality DNA template:
- Use a decent DNA extraction protocol to extract DNA, as per the sample type.
Strong Recommendation:
Use a ready-to-use spin column kit to extract the DNA.
- Assess DNA quality manually using a Nanodrop lite, Qubit fluorometer or simple agarose gel electrophoresis. Use only 1.80 (260/280 ratio) DNA.
- Use fresh DNA if possible.
- Store the DNA sample between -20°C or -80°C. This will maintain the integrity of the DNA.
- Dilute the DNA as per the sequencing requirement. Don’t use too high or low DNA concentration.
PCR amplification errors:
- Optimize primer designing to overcome primer dimer and non-specific amplification. Choose only a well-optimized and single-target primer.
- Adjust PCR condition, for instance, annealing temperature to compromise the amplification and avoid non-specific binding and primer dimer.
- Use the appropriate number of PCR cycles. I recommend using 30 to 35 cycles.
- Use high fidelity DNA polymerase. This has proofreading activities that can reduce the chances of wrong nucleotide incorporation.
- Manually investigate contaminants, primer dimers and non-specific amplification by gel electrophoresis.
Strong recommendation:
Perform post-PCR purification to remove unused dNTPs, template DNA, polymerase or other ingredients.
- Indication: Following these steps reduces the chances of background noise by 80 to 85% (from personal experience).
Dye artifacts:
- Use a proper dye concentration. I recommend referring to the manufacturer’s guide for better results.
- Use proper dye-to-template ratio.
- Monitor the incorporation of the dye to prevent excess dye usage.
- Use only high quality sequencing dye.
- To overcome between sample vs dye-based issues, run a positive control to know if the problem persists due to problems in either sample preparation or dye.
- Minimize dye exposure to light.
Incomplete termination:
- Use high quality DNA polymerase. Also, you can use high-fidelity DNA polymerase.
- Optimize sequencing conditions, for instance, increase or decrease the concentration of ddNTPs as per the background signals recorded.
- Increase (sequencing) extension time to sufficiently perform termination. This will prevent early termination.
- Run controls (positive and negative) to identify the problem related to background noise.
- Lastly, check the expiry of the sequencing kit, buffer composition and other ingredients.
A few more recommendations for troubleshooting:
If problems persist even after following these steps and troubleshooting options, arrange a technical audit with a company person.
Purify the PCR product with ethanol, this will give amazing results.
Expert’s advice is also recommended as Sanger sequencing is a complex and experience-based technique.
How can background noise affect Sanger sequencing results?
Reduced signal-to-noise ratio:
High background noise reduces signal-to-noise ratio, significantly. Meaning, it makes it difficult to distinguish between true signals and noise. Therefore, the whole analysis part becomes difficult.
False peaks:
It can also generate false peaks in the electropherogram which look similar to the true peaks. This will also confuse the bioinformaticians with base calling.
Lower sequencing quality:
Background noise decreases the overall quality of the base calling and electropherogram. This means that it also lowers the Pred score and the confidence over the final results.
Difficulties in long reads:
Although high background noise makes any analysis difficult. It becomes particularly difficult in the case of long-read sequencing. The reason is, in this particular scenario, over time, quality degrades. So overall It becomes difficult to read the fragment.
Base calling errors:
Irrespective of the expertise in the Sanger sequencing analysis, high background noise also creates errors in base calling. Meaning, that a base can be wrongly called and resultantly decrease the accuracy, reliability and trustworthiness of the sequencing.
Increase cost and time:
In the case of very high background noise, re-sequencing is required. So it increases the reporting cost and time.
Read more:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- Detailed History of DNA Sequencing
- Why Should Life Science Students Learn Sanger Sequencing? – Top 6 Reasons
Wrapping up:
Background noise is a significant negative factor in the Sanger sequencing. Varieties of reasons cause high to moderate background noise, with experience; it can be managed and/or learned to overcome.
If you face any difficulties in Sanger sequencing reading, analysis or even reducing background noise, you can contact our team. We will help you overcome these things and improve your Sanger sequencing learning.
And if you want to learn more, join our Sanger sequencing complete online course.
I hope you like this article. Do share it and subscribe to Genetic Education.
Sources:
Crossley BM, Bai J, Glaser A, Maes R, Porter E, Killian ML, Clement T, Toohey-Kurth K. Guidelines for Sanger sequencing and molecular assay monitoring. J Vet Diagn Invest. 2020 Nov;32(6):767-775. doi: 10.1177/1040638720905833. Epub 2020 Feb 18. PMID: 32070230; PMCID: PMC7649556.
Al-Shuhaib, M.B.S., Hashim, H.O. Mastering DNA chromatogram analysis in Sanger sequencing for reliable clinical analysis. J Genet Eng Biotechnol 21, 115 (2023). https://doi.org/10.1186/s43141-023-00587-6.