“A laboratory genetic technique or method to identify individuals using bodily samples such as blood, saliva, or hair is referred to as DNA fingerprinting.”
Or we can define DNA fingerprinting as,
“A DNA test employed to establish a link or relation between two persons or living organisms by analyzing their STR and VNTRs is known as DNA fingerprinting.”
DNA is our blueprint, the basis of life, encodes proteins, and regulates gene expression. It is made up of sugar, phosphate, and nitrogenous bases. DNA is located on chromosomes. The whole set of DNA or chromosomes is known as the genome.
Interestingly, there are several regions in our genome that are unique and hypervariable which differ among persons or organisms. That unique region has been applied for DNA fingerprinting.
The present technique has great utility in criminal verification and crime scene investigation. Moreover, it establishes a relationship between two persons and reveals someone’s identity.
It builds a relationship not between humans but also among any organisms that exist on earth. So it is more valuable.
Since 1984, it has been a gold-standard method for a person’s biological verification. Two persons differ biologically and have unique DNA patterns present in ‘so-called junk/useless’ genomic regions.
And henceforth, the non-coding region is important for us.
This blog post has interesting information regarding DNA fingerprinting. In the present article, I will explain what DNA fingerprinting is, its definition, process, steps, applications, advantages and disadvantages.
The article will surely add more value to your genetic knowledge and help you in your academics.
Stay tuned,
Key Topics:
What is DNA fingerprinting?
It’s a technique that observes DNA and identifies a person. It has other rare good names which are DNA profiling, DNA testing, DNA analysis, Genetic profile, DNA identification, genetic fingerprinting, and genetic analysis.
We have similar DNA sequences in all bodily parts, tissues and cells (except germ cells). It remains the same even after death. It can remain stable even after 1000 years.
99% of our DNA is similar, approximately. A 0.1% difference is sufficient to make someone so unique. The huge portion has repetitive DNA sequences and varies among organisms and even individuals.
Broadly, our genome has non-coding DNA (~97%) and coding DNA (~3%). The coding regions have genes that make proteins while the non-coding regions have transposons, pseudogenes, repetitive sequences and other uncategorized DNAs.
The DNA fingerprinting technique relies on the repetitive nature and polymorphic property of the junk DNA. Among individuals, organisms and species the number of repeats and sequence structure varies.
Researchers prepare a DNA fingerprint or a profile using that.
Now here we have to understand some basics before going further. First, let’s understand what the satellite regions of chromosomes are.
Satellites DNA:
In Genetics, the satellites are repetitive DNA regions, located on telomeres and centromeres and prevent end replication problems. Satellite regions on telomere shorten over a period of time and prevent aging.
Telomere shortening causes aging. The satellite DNA is non-coding.
We have two major types of satellite DNA present in our genome, which are either minisatellite and microsatellites.
The Minisatellite region contains repetitive DNA sequences of 10 to 60 bp. Our genome consists of 5 to 50 repeats of minisatellites. For example, VNTRs.
It has highly variable, polymorphic, unique, GC-rich sequences. As we said above, It is found mostly in telomeric regions (90% sequences).
Contrary, microsatellites are smaller than minisatellites. Microsatellites have a length of 1 to 6 bp and 5 to 10 repeats in our genome. For example, STR and SSR. Much like the VNTRs, it’s hypervariable, non-coding and telomeric.
We have covered the entire segment on VNTR and STR in the genetic markers article, for a more detailed understanding of genetic markers read the article: Genetic markers.
The first microsatellite was discovered by Jeffrey and his coworkers in 1984. The name microsatellite was given by Litt and Luty in 1989.
An interesting story:
Everyone assumes that the name satellite is given because the sequences are located on the telomeric region of chromosomes, is it! The concept isn’t true.
The technique of DNA fingerprinting changed the era of identification, characterization, and classification of organisms.
Notably, approximately 99% of our DNA (deoxyribose nucleic acid) is similar. A 0.1% difference is sufficient to make someone so unique. We are using this 0.1% portion for DNA profiling which is often known as DNA fingerprinting.

There is a reason, why only DNA is used,
In all bodily parts, tissues, and cells (except germ cells) our DNA is the same. Even, after the death of the individual, it remains the same. It can remain stable even after 1000 years.
As we know, 97% of our genome is non-coding, repetitive, and junk, protein formation is regulated by only a 3% portion, we know it as genes.
Moreover, the repetitive nature and polymorphic properties of junk DNA are utilized to perform DNA fingerprinting. The number of repeats and the sequence structure of those regions varies between individuals and organisms based on that the entire DNA profile or the DNA print can be prepared.
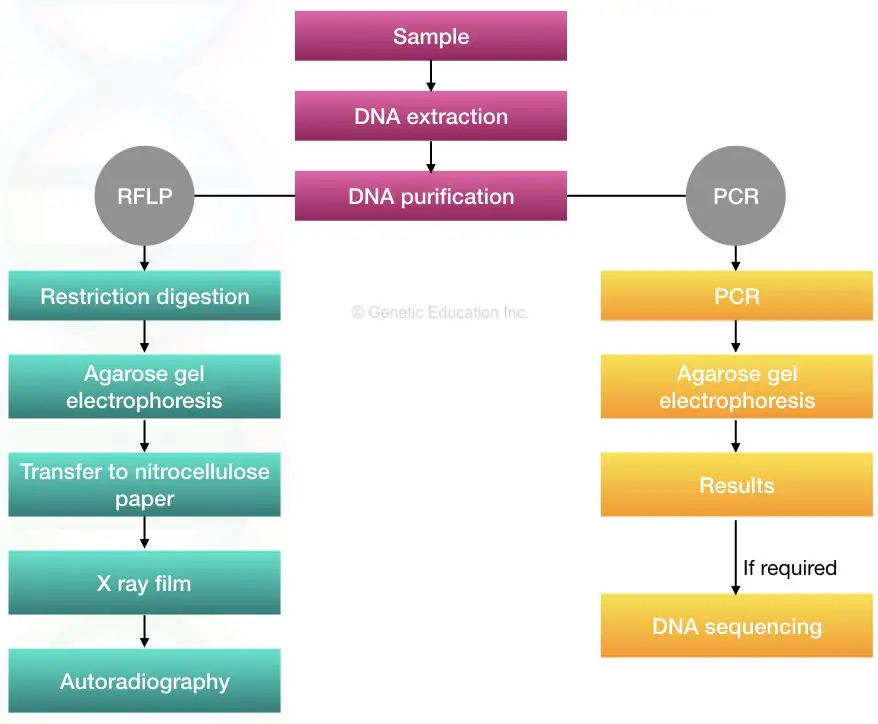
A comprehensive overview of the whole method is given below,
The DNA is isolated from the bodily sample, digested or amplified using restriction digestion or PCR, respectively. Using either agarose gel electrophoresis or sequencing, the DNA fragments are separated and identified.
Though the PCR-based method for DNA fingerprinting is accurate and too fast but after the discovery of real-time PCR and DNA sequencing, it becomes even more powerful. The molecular tools available nowadays create 100% accurate patterns of a person’s DNA.
The name ‘satellite DNA’ has been given because of its separation nature in the centrifugation process. As we said, our genome has a huge portion of repetitive sequence which appears as a thick prominent layer on the top of the test tube after the centrifugation.
That’s why it is known as satellite DNA. Now get back to our original topic. A comprehensive overview of the whole method is given below,
Isolating DNA from a biological sample following either restriction digestion or PCR amplification. Techniques to interpret results are agarose gel electrophoresis or DNA sequencing. Results show either well-separated sequences or fragments (bands) of DNA.
The PCR-based DNA fingerprinting technique has high accuracy & speed and is popular too. But the discovery of real-time PCR and DNA sequencing has changed the scenario. The technique becomes more powerful afterward.
Tools available in recent times can create a 100% accurate pattern of a person’s DNA.
Definition of DNA fingerprinting:
“Using laboratory techniques such as PCR, DNA sequencing or restriction digestion, a unique pattern of a person or any living thing can be prepared using DNA fingerprinting.”
History of DNA fingerprinting:
Since the discovery of DNA in 1953 by Watson and Crick, genetic science has become more advanced and accurate. To know more on DNA and the history of genetics, please read this article: DNA: Definition, Structure, Function, Evidence, and Types
DNA fingerprinting technique was originally developed by British scientist Alec Jeffreys in 1984. However, Dr. Lalji Singh is known as the father of DNA fingerprinting in India.
Using the restriction digestion length polymorphism, Jeffrey created the first DNA profile. His method was actually a combination of RFLP and autoradiography.
Jeffreys and coworkers identified the first microsatellite. The DNA fingerprinting was performed through RFLP & autoradiography by them.
Jeffrey had performed restriction digestion using REase and separated various DNA fragments using agarose gel electrophoresis. In the next step, the separated DNA fragments were transferred to a nylon sheet to perform southern hybridization. Radio-labeled probes were hybridized to detect various fragments.
The results were analyzed with the help of the X-ray film. This was the first method adopted by scientists to prepare a DNA fingerprint.
The first microsatellite was discovered by Jeffrey and his coworkers in 1984. The name microsatellite was given by Litt and Luty in 1989.
The technique of DNA fingerprinting changed the era of identification, characterization, and classification of organisms.
Before going further on different techniques of DNA fingerprinting, let’s first understand the correlation between tandem repeats and DNA fingerprinting.
Related article: What is DNA footprinting?- Principle, Steps, Process and Applications.
The role of a non-coding region:
Gene exists in nature because it can encode various proteins. Through the collective efforts of replication, transcription and translation; form DNA, mRNA, and protein forms, respectively.
But the DNA other than genes isn’t able to construct any protein, however, as per recent findings, non-coding DNA regulates gene expression. The loosely packed, junk and non-coding DNA regulates gene expression.
VNTRs and STRs:
“Tandem repeats are the sequences which are located one after another into the genome.”
It varies from individual to individual. For example, if a person “A” has 45 VNTRs (with 20bp) and 9 STRs (with 5bp), The possibility of having this same number of repeats for this specific VNTR and STR in another individual is almost negligible.
Nonetheless, in the case of monozygotic twins, it is quite possible.
A single embryo forms the monozygotic twins (due to the splitting of an embryo), so the chances of having the same VNTR and STR profile are higher. Still, monozygotic twins aren’t similar biologically. Read this article to know more: Do Identical Twins Have The Same DNA?
For normal individuals, the chance of having the same DNA profile is 1 in 10,000,000,000,000( the total world population is 7,600,000,000, imagine the possibility).
Difference between VNTR and STR:
|
VNTR |
STR |
| Variable number of tandem repeats | Short tandem repeats |
| A type of minisatellite | A type of microsatellite |
| Consists 10 to 60 bp | Consists 1 to 6 bp |
| 10 to 1000 repeats in a genome | 5 to 200 repeats in a genome |
| Produce heterogeneous array | Produce homogeneous array |
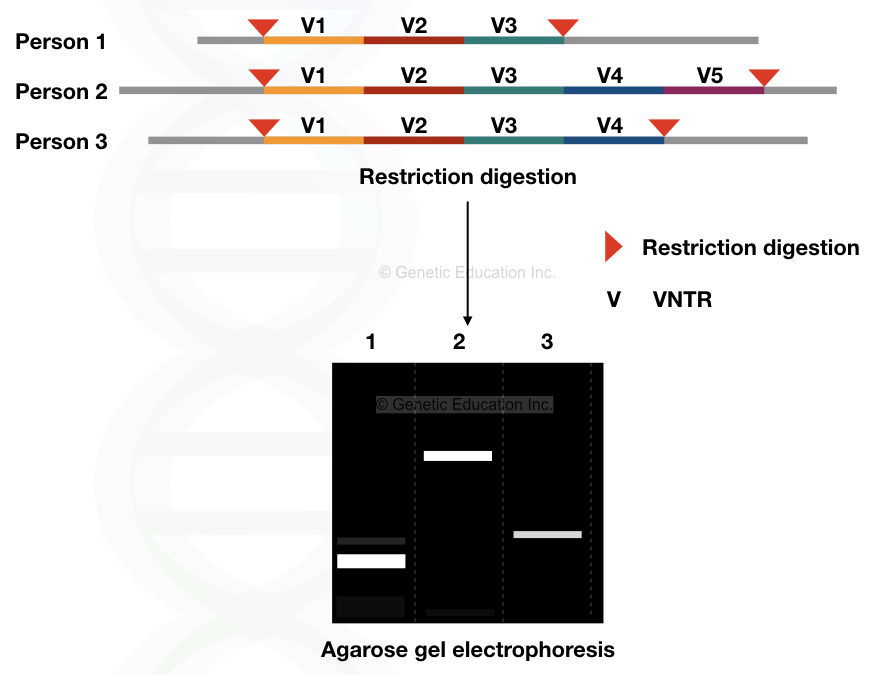
Here we have explained one simple example to understand the topic more precisely. For example, various VNTRs (V1, V2, V3, V4 and V5) are digested with a single type of restriction endonuclease, and the fragments are serrated on the agarose gel.
The figure below shows the results,
DNA fingerprinting techniques:
Step 3: Restriction digestion, amplification of DNA sequencing
Three common methods are:
- RFLP based STR analysis
- PCR based analysis
- Real-time PCR analysis
1. RFLP based STR analysis:
Restriction fragment length polymorphism was the first method used for DNA fingerprinting. Here, the restriction endonuclease- a type of nuclease cleaves DNA in various sized fragments that are separated on a gel, usually an agarose gel.
Each DNA fragment runs at different speeds due to size differences. Now the fragments are transferred to the nitrocellulose paper to perform the Southern blotting. To fix DNA on a nitrocellulose paper, chemical denaturation is performed.
In the next step, probe hybridization occurs using the radiolabeled probes. The probes find their complementary sequence and bind to it.
The probes and denatured DNA on nitrocellulose paper are now incubated for several hours to hybridize. Every time when a probe finds its complementary sequence it binds to it. Using a washing step, unbounded probes are removed. The unbounded probes complicate our results.
Now our hybridized blot is ready. It is exposed to an X-ray to get results. Different DNA bands are observed in X-ray films. The present method was widely used for DNA fingerprinting, nonetheless, it had several limitations.
It’s a tedious, time-consuming and expensive process. Furthermore, it needs more DNA therefore less recommended for forensic samples.
2. PCR based analysis:
The PCR-based DNA fingerprinting technique is the most popular among all. It is simpler than RFLP-autoradiography and gives reliable results than traditional techniques.
PCR-based DNA fingerprinting is faster and more accurate. A brief overview is given into the figure below,
Now, let’s understand the actual process.
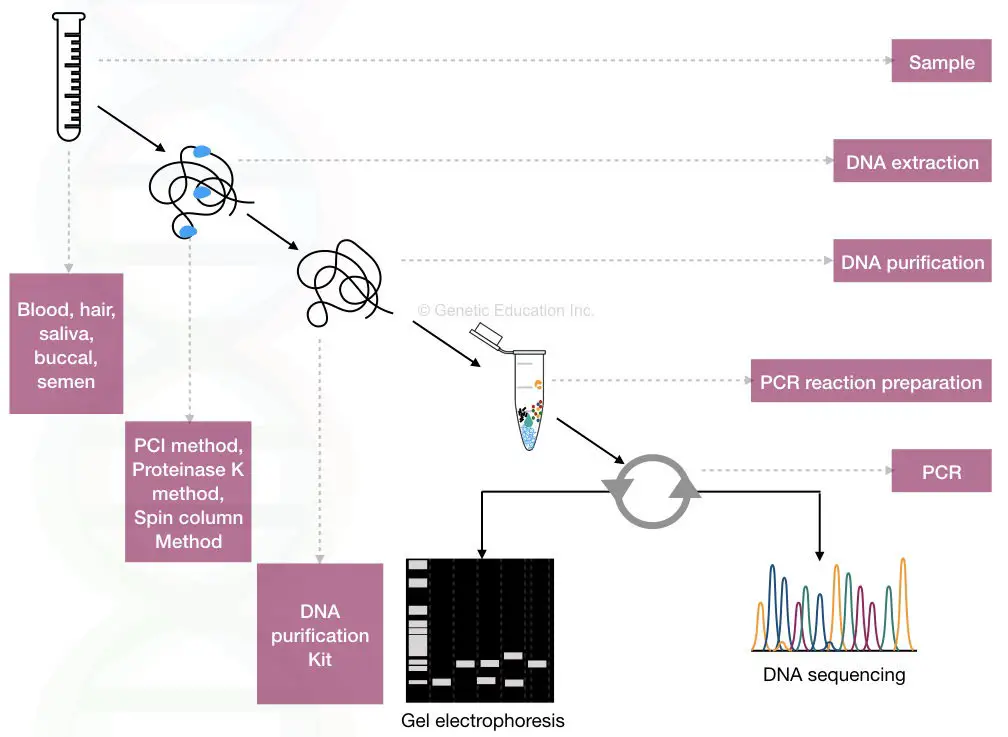
Now we have numbers of known STRs and VNTRs present in the NCBI database. We can choose any of the STR or VNTR of our interest. After choosing the sequence, primers are designed based on the sequence data.
The primer set amplifies the DNA during PCR. The steps include DNA extraction, PCR amplification and gel electrophoresis. A simple agarose gel electrophoresis setup is enough to obtain results.
Here, the amplified DNA fragments are analyzed on the agarose gel. Based on the size of DNA fragments, different DNA bands appear in a gel. However, PCR-based gel electrophoresis is mostly used for VNTR analysis. Because VNTRs are larger than STRs and separate properly.
STRs are smaller in length so it is difficult to distinguish them in an agarose gel. In contrast, VNTRs are larger fragments and give a beautiful distinct band pattern in agarose gel electrophoresis.
Nowadays, a recently evolved state-of-the-art method known as capillary electrophoresis has the potential to separate even minute DNA fragments, precisely. It can discriminate nearer fragments of single base pair difference thus for both VNTR and STR analysis, the present method is employed so often.
However, we can’t count the number of repeats present in a DNA sequence using the capillary gel electrophoresis.
3. Real-time PCR analysis:
Real-time PCR is able to amplify DNA as well as counts the number of amplicons, consequently counts the number of copies present in a sample.
The Real-time PCR can quantify the template and so is accurate. Crime scene investigation and criminal verification rely on this technique. It is quick, accurate, reliable, and cheaper than other methods. Moreover, it can even quantify a smaller amount of DNA from any sample.
The rt-PCR uses fluoro-labeled probes that hybridize with the target and give signals for quantification. Note that the probes of rt-PCR aren’t longer like the ones used in the southern blot.
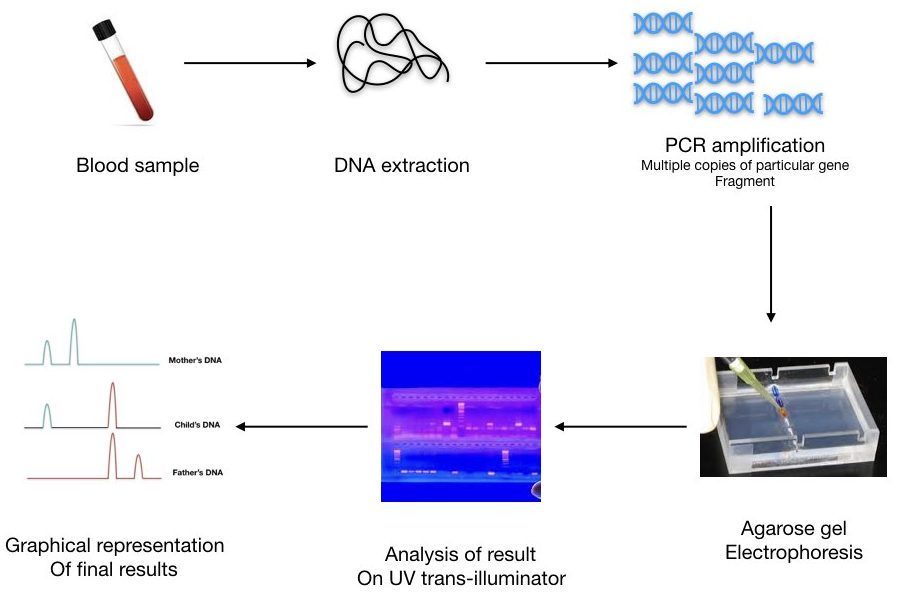
Based on the fluorescent signals obtained, it creates different peaks for different repeats. Here, gel electrophoresis isn’t required therefore this technique is so fast and very reliable.
We can calculate the number of repeats present in different samples. A pictorial representation of three different DNA bands of mother, child, and father is shown in a figure below,
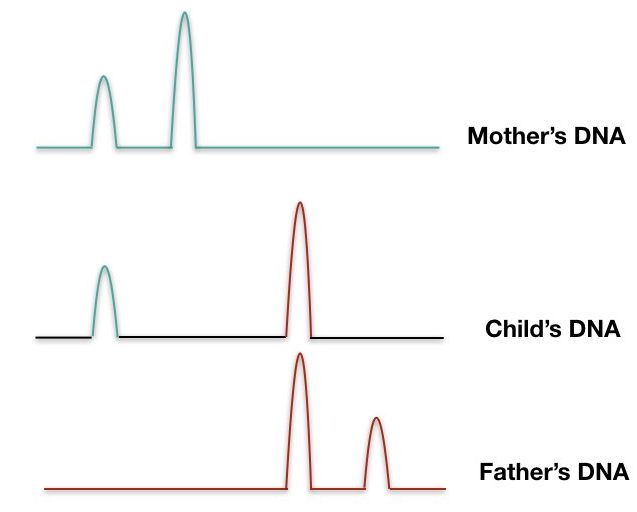
The forensic analysis uses real-time PCR along with capillary gel electrophoresis which quantifies the repeat as well as separates it too.
For other purposes, scientists use DNA sequencing to get sequence information too.
Now, let’s understand the actual process.
Steps of DNA fingerprinting:
- Collection of a biological sample- blood, saliva, buccal swab, semen, or solid tissue.
- DNA extraction
- Selecting a technique- RFLP, PCR amplification, real-time analysis or capillary gel electrophoresis
- Results analysis and interpretation.
Process of DNA fingerprinting:
Sample collection, DNA extraction, digestion or amplification and analysis results are major steps.
Step 1: Sample collection:
We can get DNA from any bodily sample or fluid. Buccal smear, saliva, blood, amniotic fluid, chorionic villi, skin, hair, body fluid, and other tissues are the major types of samples used.
In criminal cases, a buccal swab is usually taken. The buccal swab sample collection method is non-invasive and handy.
However, if not maintained properly, it can easily be contaminated with bacteria. Further, the Buccal swab DNA yield is very less. A blood sample is a good replacement for a buccal swab sample. We can use a blood sample as well.
Step 2: DNA extraction
We have to first obtain DNA.
To perform any genetic applications, DNA extraction is one of the most significant steps. Good quality and quantity DNA increases the possibilities of getting good results.
You can use either of the DNA extraction methods listed below,
Nevertheless, we strongly recommend using a ready-to-use DNA extraction kit for DNA fingerprinting.
The purity and quantity of DNA should be ~1.80 and >100ng, respectively to perform the DNA test. Purify the DNA using the DNA purification kit, if needed.
After that, quantify the DNA using the UV-Visible spectrophotometer. And perform one of the following methods listed below,
Step 3: Selecting a technique:
What technique to use depends on what assay you are performing, for instance, PCR-based gel electrophoresis is sufficient for detecting maternal cell contamination.
On the other hand, capillary gel electrophoresis and real-time PCR is employed for forensic analysis.
Each technique has its importance and limitations. Here is the list of advantages and disadvantages of each technique.
| Technique | Advantages | disadvantages |
| RFLP | Accurate and cheap | Tedious and time-consuming process.
Need more DNA samples. |
| PCR-agarose gel electrophoresis | Quick, easy to use, cheaper, accurate and reliable | Can’t count the number or repeats present.
Can’t distinguish smaller DNA fragments. Can’t give sequence information. |
| Rt-PCR | Fast, easy to use, cheap, accurate and measures the number of repeats. | Can’t distinguish smaller fragments.
Can’t give sequence information. |
| Capillary electrophoresis | Distinguish smaller DNA fragments up to 1 bp change. | Can’t count the number of repeats present. |
| DNA sequencing | Provides sequence information. | Time-consuming and costly. |
Step 4: result and interpretation:
As we discussed, technique to technique, the results vay and the analysis process too. Here is the summary.
| Technique | Results |
| RFLP | Autoradiography- DNA bands are observed on a film. |
| PCR | Agarose gel electrophoresis- DNA bands are seen on a gel |
| rt-PCR | The amplification curve shows different amplicons present in a sample |
| DNA sequencing | The sequence information of the target |
| Capillary gel electrophoresis | Separates even minute bands and distinguishes them. |
By comparing the DNA profiles of various samples, information of variations and similarities between individuals can be obtained. Notably, the entire process is now almost automated. We do not have to do anything, the computer gives us the final results.
Applications of DNA fingerprinting:
The revolutionary DNA fingerprinting technique has many applications in diverse fields from forensic to medical science. Here in the section, I am enlisting several popular applications of DNA fingerprinting.
Revealing a person’s identity.
Biological identity is the 100% unique thing for a person, in fact for any organism on earth. DNA analysis makes it possible to reveal someone’s biological identity. Perhaps it’s the only better option available.
Identifying dead bodies.
Yes, you hear it right! The present technique can even identify dead bodies when mass disasters occur. Meaning, it can identify a dead body based on available data of a person.
Again, it is almost 100% accurately identify someone, biologically. The technique helps to identify badly damaged dead bodies.
Identification of blood relatives.
No two individuals are genetically identical, we know that. To establish or to know the blood relationship between two unrelated individuals, the present method has been adopted.
The examiner collects biological samples from two individuals, parents or children, extracts DNA, performs a test and examines results. Results conclude whether the samples belong to blood relatives or not.
Crime scene investigation and criminal verification.
One of the most important applications of the present technique is in crime scene investigation and criminal verification.
Police or examiner collects samples from the crime scene, the sample might be saliva, blood, hair follicles, semen or anything that is sent for fingerprinting.
An expert processes sample using two marker systems (explained above) and analyze them against the suspect. Results establish a link between criminal and sample obtained from the crime scene.
Different countries have different criteria to use STRs. The US, UK and India use 13, 11 and 12 STR markers to investigate crime. The more the number of markers, the more accurate results are.
Organ transplantation studies.
One of the important applications of DNA profiling is in checking graft acceptance and rejection in the case of organ transplantation. The HLA typing technique has been employed for transplantation studies.
Our body naturally resists or tries to destroy foreign antigens, pathogens, particles, molecules or even organs. Therefore it’s essential to check whether donor’s and recipient’s HLA types are matching or not.
HLA types are Human Leukocyte Antigens involved in the immune system. Various HLA loci are examined for graft and transplantation study. The exact match score indicates graft acceptance. This means researchers can transplant the organ.
Maternal cell contamination detection.
One of the best uses of DNA profiling/fingerprinting which I know, probably others don’t know, is detecting MCC.
Prenatal studies and diagnosis rely on using chorionic villi samples and amniotic fluid. It sometimes consists of maternal cells too, especially when isolating CV samples.
Meaning, contaminated tissues also have maternal cells which mislead the results of a test. It produces false-positive results. It becomes more vulnerable when it comes to heterozygous or carrier identification.
Using a simple PCR-agarose gel setup, taking a couple of VNTR and STR makers, the maternal cell contamination in the original prenatal sample can be identified. It makes pregnancy genetic testing successful and 100% authentic.
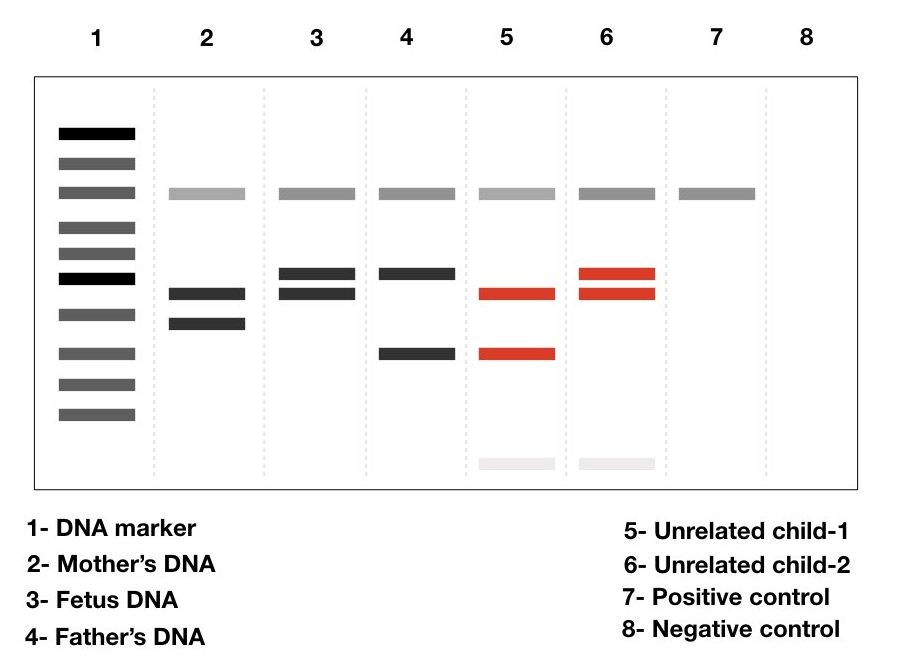
Studying genetic disease.
The best application of DNA fingerprinting has in medical science, to study genetic diseases. Scientists use various modifications of PCR to examine and study genetic diseases.
Mutations and allele variations are also studied too. It can also create phylogeny between various organisms.
My experience of DNA fingerprinting
During my tenure of prenatal research, instead of following the standard protocol with STR or VNTR, I had prepared my own combination of markers.
I had selected 4 VNTRs, 3 STR, 1 mitochondrial DNA marker and 1 Y chromosome-specific STR for the detection of maternal cell contamination.
Mitochondrial DNA is additionally a great tool for verifying individuals. The number of mitochondrial DNA varies between cells, individuals and organisms. The Y-specific marker helps in gender verification.
Let me give you a spoiler! The forensic analysis includes one mitochondrial marker and a Y-specific marker.
Conclusion:
Scientists have been using DNA fingerprinting for many different applications for a long time. However, it has become more popular in crime scene investigation and settling parental identification claims.
The technique is accurate, reliable, fast, and cheap though changes may occur timely. Traditionally, researchers have relied on restriction digestion, but PCR has made it more aggressive.



I have a doubt sir,. By using fingernail scraping as a sample,we can run the STR gel electrophoresis or cant?
No, fingernails are made up of protein.
This is what I needed for my project on DNA Fingerprinting.Thanks a lot Sir.
Thank you abdur Rahman
Such a lovely one it helped me alot in writing assignment..it just amazing..thnx alot
Thank you Prajwala Harmalkar
Thank you sir
Excellent explanation and very easy to understand. Helped me a lot thanks.
Thank you Sanjana Jain
excellent information and such a brilliant way of conveying the whole lecture.
Thank you sultan