“The Sanger sequencing technique is a gold-standard method for DNA sequencing and is widely used routinely in various fields.”
Sequencing techniques allow scientists to determine or ready nucleotides from any DNA fragment or gene. This eventually provides data regarding mutations and alterations within the sequence.
Among the first, second and third generations of sequencing, Maxam Gilbert sequencing and Sanger sequencing are two platforms, that belong to the first-generation sequencing. These techniques are time-consuming, slow and error-prone.
However, due to the use of hazardous chemicals and high error rate, the Maxam Gilbert sequencing did not gain popularity like the Sanger sequencing. The technique developed by Sanger and co-workers evolved as technology advances.
It adopted automation and became a popular and gold-standard method in genetics and clinical diagnostics. In this article, we will witness the journey of Sanger sequencing from a traditional gel-based approach to a fully automated capillary approach.
Stay tuned.
Key Topics:
What is Sanger sequencing?
In 1977, Frederick Sanger and his team developed a novel sequencing approach. In this approach, using a unique ingredient in four separate reactions can help to determine the order of nucleotides from a sequencing.
During that era, Sanger sequencing highly relied on the PAGE-based separation approaches, the reaction was run on a gel to determine the fragment size and nucleotide orders.
Confusing!
Explore from the beginning.
The Sanger sequencing reaction is similar to the PCR one! In PCR, we use dNTPs, Taq DNA polymerase, amplification buffer, primers, template DNA and water to amplify the target DNA.
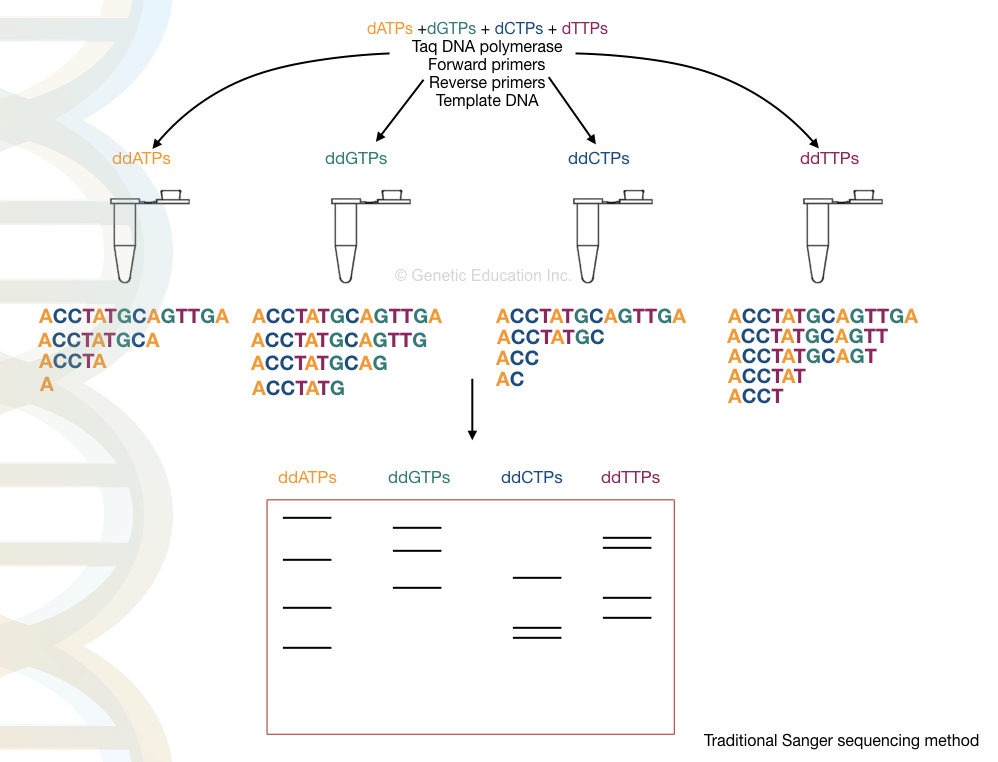
Sanger sequencing approach uses the same ingredients with an additional ingredient that is— ddNTPs. The above-listed ingredient synthesizes the target DNA, however, when it encounters the ddNTP in the Sanger sequencing, the chain synthesis is terminated.
Do you know why?
The DNA polymerase needs a primer and a 3’-OH end for initiating DNA amplification, which is provided by the primer set we use in the PCR. The DNA polymerase uses a free complementary dNTP and continues DNA amplification.
ddNTPs are a special class of nucleotide base or base analog that doesn’t have the free 3’-OH group which means when the polymerase encounters a complementary ddNTP, it terminates chain synthesis.
To know more, you can read our comprehensive article on this concept: ddNTPs in Sanger Sequencing- what it is and why to use it. Thus, the process is also known as chain termination.
These fragments are then run on the PAGE gel to investigate each terminated fragment.
Principle of Sanger sequencing
Sanger sequencing relies on the principle of chain termination. That means, in the four different reactions, when the polymerase encounters the ddNTP, it terminates the chain synthesis process.
All four different reactions are run in a PAGE gel. Fragments are read from smallest to largest, in the gel to determine the nucleotide sequence.
Process:
The Sanger sequencing process has been divided into four different steps: DNA extraction, PCR amplification, electrophoresis and results analysis.
DNA extraction:
A high-quality genomic DNA from the sample is isolated using a well-standardized protocol, a kit or an automated system. The DNA should have a purify of 1.80 (260/280 ration), and quantity >50nG (in general) for the sequencing.
However, experts can also perform sequencing even with a very small volume of DNA.
PCR amplification:
Now, the target fragment is amplified using a PCR protocol. The goal here is to generate amplicons for the desired gene of a fragment. The amplicons can be validated using a conventional agarose gel electrophoresis.
A standard primer design guidelines, reaction preparation, PCR protocol and amplification conditions are used. Note that PCR purification is mandatory to remove non-used dNTPs, Taq DNA polymerase, primers and other chemicals before preparing the sequencing reaction.
Sequencing:
Now the sequencing reaction is prepared using the fluorescent-labeled ddNTPs along with all the essential amplification ingredients. As we only need a single sequence, a single primer either forward or reverse is needed here.
After completion of the sequencing run, the sequencing product is first purified. For instance, ExoSAP-IT is a purification kit available from ThermoFisher for sequencing reaction purification.
Unused ddNTPs may fluorescence during the fragment analysis and give background signals that’s why purification is needed.
Capillary electrophoresis:
Now, the fragments (sequencing product) are run on the capillary electrophoresis, this will separate fragments based on the size. After run completion, it generates a sequencing file that contains fluorescent peak, peak quality and sequence information.
In the next step, the file is analyzed using software.
Results and analysis:
Many software are available for Sanger sequencing analysis. The file can be downloaded and analyzed in any of the software. After analysis, the sequencing file is generated that can be used for BLAST analysis.
BLAST compares the nucleotides of the sequence file with the reference data to identify nucleotide alterations.
Traditional Sanger sequencing
The traditional Sanger sequencing approach was an original procedure developed by Frederick Sanger. ddNTPs, chain termination and PAGE-based analysis are three unique approaches used in the original process.
Radiolabeling was adopted in the initial Sanger sequencing approach to investigate fragments after the gel electrophoresis. The DNA fragments are labeled with either P32 and S35 to study them using autoradiography.
In four different tubes, four different chain termination reactions for each ddNTP (ddATP, ddGTP, ddCTP and ddGTP) are performed.
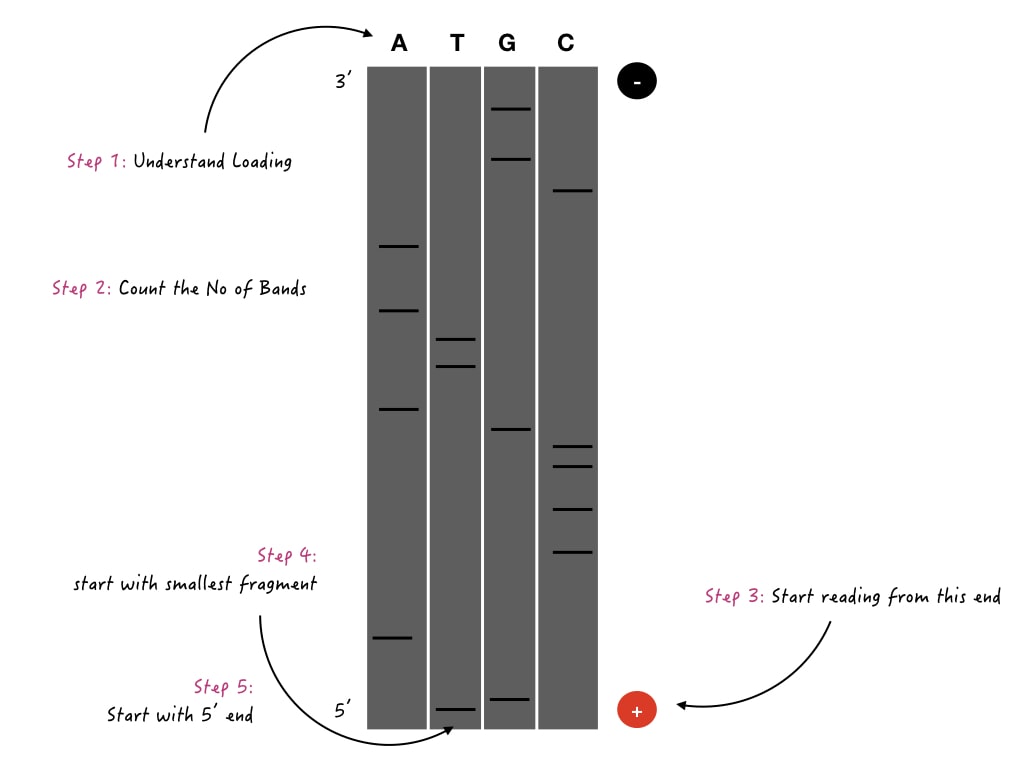
A PAGE gel (Polyacrylamide gel) is prepared and run on a vertical gel aperture. Four different reactions are loaded in four different wells and allowed to run for a few hours. PAGE can differentiate smaller DNA fragments effectively.
After completion of the electrophoresis, the gel is placed on the photographic plate, the radiation emitted from each fragment is then captured and a visible gel bands’ image is created.
Scientists invest hours to study the gel image and correctly locate each nucleotide in the sequence.
Traditional Sanger sequencing is required,
- Radiolabeling, which is hazardous for the lab personnel.
- Tedious sample preparation.
- Four different reactions to setup.
- Tedious gel preparation and setup.
- Autoradiography for image analysis.
- Human interference for results and analysis.
Thus the whole process is tedious, time-consuming, error-prone and labor-intensive. If the investigator makes a mistake in reading the band, the entire sequence information can be misinterpreted.
Nonetheless, the traditional approach remained popular for many years as it was handly and streamlined, at that time.
Automated Sanger sequencing:
Semi-automated and fully automated approaches were developed after 1985. Applied Biosystems (ABI) made a first attempt to replace the traditional Sanger sequencing approach with a semi-automated approach.
They first, replaced the radiolabeling with the fluorescent labeling. However, the core concept of chain termination remained similar in the automation as well, but with a slight modification.
Substantial, automation was made after the introduction of capillary gel electrophoresis. EC uses smaller capillaries to run the fragments instead of a gel. The capillaries are so small that they can even distinguish a single base-pair change.
The integration of fluorescent ddNTPs and CE in the Sanger sequencing changed the entire sequencing industry. Here, all four ddNTPs that are responsible for chain termination, are labeled with different fluorescent colored tags.
Only a single reaction is prepared in which all the four “labeled” ddNTPs are added along with the standard amplification ingredients. On the other side, at the end of capillary gel electrophoresis, there is a laser.
Now, after completion of the chain termination sequencing reaction, the single sequencing reaction is run in the capillary electrophoresis unit. All the fragments are run based on their size in the capillary.
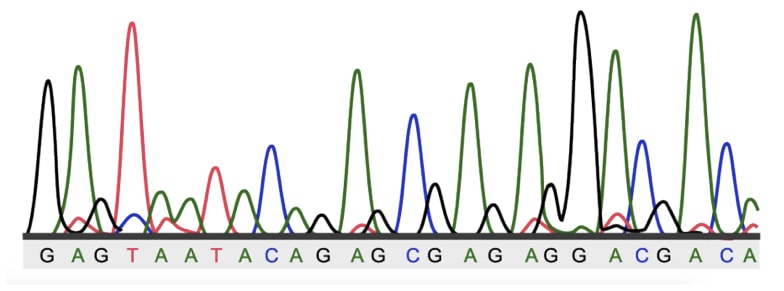
Now when it reaches the edge, the laser light is when strikes the fluorescent tag, the fluorescence is released and reported. The computer software prepares a fluorescent peak based on the intensity and color, and reads the nucleotide.
This is the automated Sanger sequencing process. In recent times, DNA extraction can also be done using an automated extraction unit, which makes the entire procedure even more streamlined and automated.
Automated Sanger sequencing required,
- Fluorescently labeled ddNTPS, which are safe to use.
- A single sequencing reaction that saves time and reagents.
- Capillary electrophoresis- which doesn’t need any gel preparation and also has the power to distinguish even a single base pair change.
- Computer software, that doesn’t need any human interference.
Thus, the automated Sanger sequencing approach is fast, more accurate, easy, time-saving and safe.
Traditional vs Automated Sanger Sequencing
| Traditional Sanger sequencing | Automated Sanger sequencing | |
| Method | Chain termination with ddNTPs | Chain termination with ddNTPs |
| DNA extraction | Manual methods | Ready-to-use kits and automated extraction |
| Sequencing reaction | Four different reactions for four different ddNTPs. | A single reaction for all ddNTPs. |
| Labeling | Radiolabeling of DNA fragments | Fluorescent labeling of ddNTPs |
| Separation technique | PAGE- Polyacrylamide gel electrophoresis. | CE- capillary gel electrophoresis |
| Analysis | Autoradigraphy with manual investigation. | Computational analysis using software that doesn’t require human interference. |
| Error rate | Very high | Low |
| Throughput | Low | High |
| Time efficiency | Time-consuming | Time-saving |
| Accuracy | Low | High |
| Safety | Hazardous | Safe |
| Read length | Small (500 to 600bp) | Large (800-1200bp) |
Related article: Sanger Sequencing vs PCR: Common and Technical Differences.
Wrapping up:
The automated Sanger sequencing method is a gold-standard method in the field of research and clinical diagnostics. However, it was indeed developed from the manual method.
The present sequencing approach is used to diagnose genetic conditions and study genes and DNA sequences. Furthermore, it is widely used in 16s rRNA gene sequencing for microbial typing.
I hope you like this article.