“Direct PCR is a technique that allows direct sample amplification, without DNA extraction, saving cost and time and improving outputs. Explore this comprehensive guide on direct PCR.”
What’s the most tedious part of your research project that challenges your dissertation? The extraction part!
DNA extraction is a tedious, time-consuming and error-prone process, which directly impacts the PCR amplification. Students have to invest a huge part of their dissertation only to extract the DNA.
And if I am not wrong! Oftentimes, they have to re-extract DNA for a few samples. However, amplification is still not guaranteed.
What if I tell you, one technique helps you to overcome this limitation? You don’t need to perform DNA extraction. Take a sample, perform amplification; that’s it! And, this technique is also popular in forensic.
I’m talking about the direct PCR.
Let me introduce you to the concept of direct PCR. In this article, we will cover the concept, sample types, procedure, protocol, advantages, limitations and applications of direct PCR.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy to understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Key Topics:
What is Direct PCR?
Direct PCR has existed since 1990!
Direct PCR is an amplification technique used to amplify or quantify (using qPCR) the DNA directly from a biological sample. It allows amplification directly from any biological tissues, such as animal, plant or bacterial cells, cell culture, body fluids or solid tissues.
It’s a simple, fast and effective way to perform an amplification. The DNA can be directly amplified in the presence of contaminants or impurities, and usually requires a smaller amount of DNA.
Two ways to do direct PCR are using the direct protocol and the dilution protocol. We will discuss that later in this article. Now we will look for why we need it.
Why do we need direct PCR?
~83% of DNA can be lost during the extraction procedure, which is even higher when using the manual techniques. Generally, DNA extraction is a time-consuming, lengthy and error-prone technique.
PCR requires highly pure DNA. Our cells usually contain various PCR inhibitors that first need to be removed. Moreover, the DNA should also be sufficient to perform the amplification.
Thus, it requires an additional DNA purification and quantification step to obtain pure DNA and quantify it for downstream analysis. In addition, extraction protocols also vary among different samples, and don’t guarantee success.
An efficient lysis procedure must be followed to remove impurities, contaminants and PCR inhibitors during extraction. And if this is not enough, the chemical used during DNA extraction or in the lysis buffer can inhibit the Taq DNA polymerase or amplification reaction in any way.
Direct PCR helps overcome these challenges and provides a way to perform better amplification. Various kits, reagents and enzymes are now available to perform direct PCR amplification.
We will discuss that later in this article.
How it works:
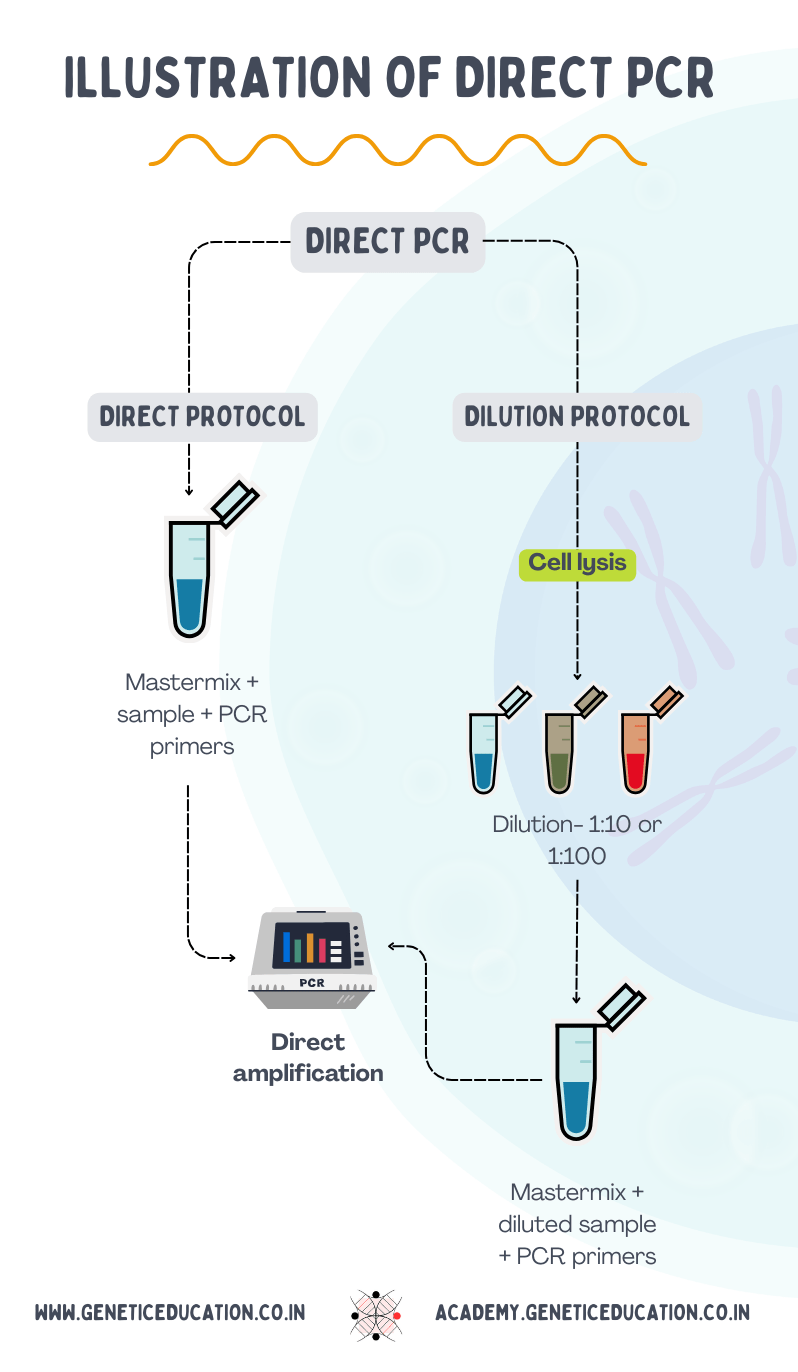
There are two ways to perform direct PCR: direct amplification protocol and dilution protocol.
In the direct protocol, a sample is directly added to the reaction mixture containing all the ingredients for the amplification. Here, a special type of DNA polymerase enzyme is used, known as inhibitor-tolerant polymerase, working even in the presence of inhibitors like proteins or polysaccharides.
The direct protocol is ideally suitable for pathogen amplification. The reason is the smooth and very fragile cell wall and the least complex cellular structure. By adding a heating step in the PCR, the DNA is released and amplified simultaneously.
It is used for TB PCR and other pathogen amplification. Keep in mind that you can prepare your own PCR mastermix and use it, but the kit will provide better results. Thus, it has been used for pathogen genotyping and colony PCR.
A dilution protocol is highly recommended for difficult tissues like animal or plant cells that contain a difficult cellular composition. The crude lysate, prepared using the lysis buffer, is diluted in a ratio of 1:10 or 1:100 before use to reduce the concentration of PCR inhibitors.
In conclusion, the direct PCR process is simple:
- Prepare the crude lysate (if needed)
- Prepare the PCR reaction
- Add the sample or lysate to the reaction
- Run the amplification protocol
| Direct PCR protocol | Dilution protocol | |
| Sample types | Whole cells, crude lysates, blood, tissue punch, bacterial colonies | Whole cells, crude lysates, blood, or swab samples |
| Lysis step | optional | Required (initial lysis, followed by dilution) |
| Incubation | Short (5–15 min at 55–65°C depending on protocol) | Short (5–15 min at 55–65°C depending on protocol) |
| Dilution | Not required | Required: 1:10 to 1:100 |
| Template volume | 1 to 2µL of the sample | 1 to 2µL of the lysate |
| Polymerase | Specialized inhibitor-tolerant polymerase | Normal Taq DNA polymerase works, but recommended to use a special one. |
Samples for Direct PCR:
Any biological sample can be used for direct amplification. However, several samples are difficult to amplify due to a higher amount of PCR inhibitors, contaminants and impurities.
Samples like bacterial cells, colonies, pathogen samples, yeast cells, mouse tail snips, insect legs or wings, buccal swabs, dried blood spots, saliva and hair follicles have a low level of PCR inhibitors and thus can be processed using the direct amplification protocol.
Keep in mind that it needs a special inhibitor-resistant DNA polymerase or kit.
Conversely, Several samples have higher contaminant and inhibitor levels and need an additional chemical or heating lysis step. These samples and their required treatment are explained here.
- Whole blood (liquid form) → Lysis and Dilution recommended
- Fresh plant tissue (leaf disc, seedling) → Requires lysis buffer treatment
- Animal tissues (e.g., liver, kidney) → Requires mild lysis or heat
- Cultured mammalian cells → Lysis buffer or freeze–thaw recommended
- Fungal mycelium → Boiling or enzymatic lysis first
- Saliva (large volume) → Dilution recommended
Note: The Direct PCR technique is extensively used in forensic analysis due to minimal sample availability.
DNA polymerase for Direct PCR
DNA polymerase synthesizes the template DNA in the PCR; therefore is crucial for PCR. However, unlike the conventional PCR, the Taq DNA polymerase can’t work efficiently for direct PCR.
Various PCR inhibitors, like proteins, polysaccharides or other components present in the sample, deactivate it. To use a polymerase in the present assay, it must have some ideal conditions.
- It must be inhibitor-resistant. It can tolerate common PCR inhibitors like proteins, hemoglobin, heparin, bile salt, polysaccharides, etc. If you want to learn more about PCR inhibitors, you can read our previous article.
- It must have high polymerization activity and a moderate to high fidelity to avoid mutation-free amplification.
- It must have a hot-start activity to avoid early amplification, non-specific amplification and primer-dimer.
- It must have high specificity for partially degraded or unpurified DNA.
- It should work for a variety of samples like blood, saliva, plant or bacterial cells/culture.
Several commercially available polymerases for direct PCR are summarised here in the table.
| DNA polymerase for direct PCR | Inhibitor resistance | Detergent tolerance |
| Phusion | Moderate | Moderate |
| KOD FX | High | High |
| BIOTAQ | High | Moderate |
| Mighty Amp | Moderate | Moderate |
| KAPA Blood | Moderate | Moderate |
*Source: Miura M.
Protocols for Direct PCR:
Here I am explaining two universal protocols for direct PCR. However, minor modifications and kit selection play a major role.
Direct PCR Protocol:
Pick a small part of tissue (e.g., a 1 mm² leaf punch) or a bacterial colony with a sterile tip.
Add the sample directly to the PCR tube containing the PCR master mix.
Complete the PCR reaction by adding
- 10µL Direct PCR master mix
- 1µL forward primer
- 1µL reverse primer
- 1µL DNA polymerase (5U)
- 12µL Sterile nuclease-free water
Prepare the 25µL reaction.
Perform the PCR using standard run conditions for 35 cycles.
- Initial denaturation: 98°C for 5 min
- Denaturation: 98°C for 5 sec
- Annealing: 55–60°C for 5 sec
- Extension: 72°C for 20–30 sec
- Final Extension: 72°C for 1 min
Run the sample on a 2% agarose gel using 5µL amplicon.
Note: a few mastermix also contain dye for gel electrophoresis, so you don’t need to purchase or use the dye separately.
Dilution Direct PCR protocol:
Lyse the sample first using 50–100 µL of simple lysis buffer.
Heat at 95°C for 10 minutes to break cells and release DNA.
Spin briefly to settle debris.
Perform sample dilution using nuclease-free water (1:10 or 1:50).
Prepare the PCR reaction:
- 10µL Direct PCR master mix
- 1µL forward primer
- 1µL reverse primer
- 1µL DNA polymerase (5U)
- 12µL Sterile nuclease-free water
Set the PCR run using the standard conditions given above.
Run the sample on the 2% agarose gel using the standard gel electrophoresis protocol.
Conventional PCR vs Direct PCR
This article is comprehensive and lengthy, so we will discuss the differences between the standard PCR and direct PCR in the form of a table.
| Conventional PCR | Direct PCR |
| Requires a well-standardized DNA extraction setup and protocol | Does not require DNA extraction |
| Needs additional purification and quantification steps | No purification or quantification required |
| Time-consuming (due to extraction and amplification steps) | Faster (amplification only) |
| Tedious and involves multiple steps | Simple and streamlined |
| Requires extraction kits, chemicals, and lab utilities | Needs only PCR buffer, mastermix, and a special enzyme |
| Higher risk of contamination due to multiple handling steps | Minimal contamination risk with fewer handling steps |
| Uses standard Taq or high-fidelity polymerase | Requires inhibitor-resistant polymerase |
| Suitable for any sample with extracted DNA | Limited to samples compatible with direct amplification |
| Generally consistent and reliable yield | Yield may vary depending on sample inhibitors |
| Used for DNA testing, diagnostics, and sequencing | Suitable for rapid screening, genotyping, and research |
Advantages:
- Simple, fast, effective and efficient.
- Requires a very low amount of DNA.
- No need for DNA extraction, purification or quantification.
- Reduce contamination risk.
- Least prone to PCR inhibitors.
- Save cost and time.
- Work on any tissues.
- High sensitivity.
Limitations:
- Not suitable for all samples, tissues or materials.
- High levels of PCR inhibitors yield inconsistent results.
- Requires a special PCR inhibitor-resistant DNA polymerase.
- Results are inconsistent due to variable DNA concentration in the crude or sample.
- Amplicons are not suitable for gene cloning or sequencing.
- Requires sample-specific optimizations.
- Due to these limitations, it is not advisable for diagnostics.
Applications of Direct PCR:
Despite having limitations, the present type of PCR is widely used in various assays and fields. Here are some of the important applications of direct PCR.
Forensic and DNA typing:
The direct PCR assay is extensively employed in forensics and DNA typing. It enables the processing of minimal samples and provides efficient and reliable results.
Colony PCR:
One of the major applications of direct PCR is in colony PCR, where the bacterial colonies are picked and employed for direct amplification. The PCR has been performed directly either on the slide or in the PCR tube.
Pathogen detection:
Yet another crucial application is in pathogen detection from environmental, veterinary and clinical samples. The simple and least complex pathogen cellular structure allows rapid amplification without requiring extraction.
Animal and insect research:
Direct PCR is extensively used in animal and model organism research, where samples like mouse tail snips or other tissues are used for rapid amplification and study.
It is also applied for insect research, where various insect body parts are directly used for amplification.
Plant and seed research:
The present technique is the least important but is still used in plant and seed research. DNA can be directly amplified from leaf, seed or root samples.
Genotyping:
A few assays and kits are now available for genotyping, allowing scientists quick genetic screening or SNP studies.
Other fields:
Besides, direct PCR is also used in genetic research, cell line validation and microbial research. Scientists are optimizing it for various other applications. However, limited kits and options are available.
Wrapping up:
Direct PCR is highly advantageous, despite having limited sample types. Scientists are developing novel kits and assays to utilize them in diagnostics, aiming to reduce the TAT and improve patient outcomes.
This article is written for students who want to optimize their research and those designing their dissertations. Choosing direct PCR not only saves time and consumables for you but also develops a novel assay for testing.
You can register or file IPR for your direct PCR assay and earn royalties! I hope you like this article. Do share this article and subscribe to Genetic Education.
Sources:
Miura M, Tanigawa C, Fujii Y, Kaneko S. Comparison of six commercially-available DNA polymerases for direct PCR. Rev Inst Med Trop Sao Paulo. 2013 Nov-Dec;55(6):401-6. doi: 10.1590/S0036-46652013000600005. PMID: 24213192; PMCID: PMC4105087.
Cascella, R., Strafella, C., Ragazzo, M. et al. Direct PCR: a new pharmacogenetic approach for the inexpensive testing of HLA-B*57:01. Pharmacogenomics J 15, 196–200 (2015). https://doi.org/10.1038/tpj.2014.48.
Song F, Kuehl JV, Chandran A, Arkin AP, 2021. A Simple, Cost-Effective, and Automation-Friendly Direct PCR Approach for Bacterial Community Analysis. mSystems 6:10.1128/msystems.00224-21: https://doi.org/10.1128/msystems.00224-21.
Martin B, Linacre A. Direct PCR: A review of use and limitations. Sci Justice. 2020;60(4):303-310. doi:10.1016/j.scijus.2020.04.003.