“Proteinase K DNA extraction method is widely used in genetic research. In this article, Learn the basics of proteinase K DNA extraction with a standard protocol.”
DNA extraction is a primary and vital step in any genetic research experiment. Starting from the very basic PCR to advanced microarray and NGS experiments, the success of any technique highly relies on how effectively we extract the DNA.
DNA extraction or isolation is a process to isolate the DNA from the cell by removing cell components including the proteins and protein composite. Proteinase K effectively digests the protein part of the cell.
The objective here is to collect a pure (260/280 nm ratio of 1.80) and high quantity (>100μg) genomic DNA for the experiment. To do so, various DNA isolation methods and kits are available.
Phenol chloroform, CTAB and Proteinase K DNA extraction methods are the three most popular and widely used manual DNA extraction methods. Among these, Proteinase K is the most accurate and trustable method.
This article contains all the essential information on the Proteinase K DNA extraction method. With this, I will also share our own standardized protocol.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is Proteinase K?
Proteinase K is an enzyme widely used in various biological experiments.
Proteinase K
Protein (long chain of amino acid) + ase (suffix for enzyme) + K (keratin). Why keratin? Here is an interesting story!
Protease enzyme was first isolated from the fungus Tritirachium album. It can effectively digest keratin. Keratin is a hair protein and the hardest form of any protein. The suffix “K” was given from its present unique characteristic.
| Name | Proteinase K |
| What it is | Broad-spectrum serine protease |
| Isolation | Derived from the fungus Tritirachium album |
| Molecular Weight | Approximately 28.9 kDa |
| Storage Condition | Store at -20°C or below, preferably in aliquots |
| Application | DNA and RNA extraction, protein digestion, inactivation of nucleases, removal of contaminants |
| Enzyme activity | Active over a wide range of pH (optimum pH 7.5–9.0) |
| Substrate specificity | Cleaves peptide bonds at the carboxyl side of hydrophobic, aliphatic, or aromatic amino acids |
| Maximum activity at pH | pH 8.0 (Range 4.0–10.0) |
| Maximum activity temperature | 65°C (Range 37 to 70°C) |
Proteinase K belongs to the family subtilisin, a typical proteinase family and a member of serine proteinase. It has a smaller molecular weight of approximately 28.9KDa. The single polypeptide chain consists of 278 amino acids.
Much like other enzymes, Proteinase K also needs a cofactor, not for activation but for functional stability. One important property of Proteinase K is its resistance against heat and chemicals.
Therefore, it is widely used in nucleic acid extraction and other biological experiments. Notedly, The first proteinase K was isolated in the year 1974 and it is the most commonly used endopeptidase; proteinase in DNA extraction.
Why is Proteinase K used in DNA Extraction?
Proteinase K is widely used in nucleic acid (DNA/RNA) extraction experiments. A cell typically consists of various forms of proteins. For example cell wall receptors, composite cell membrane proteins (glycoprotein, phosphoprotein, etc), nucleases and nucleic acid-associated proteinase.
Here is the list of various proteins and protein composition present in a cell and cell components.
| Cell Component | Protein Associated |
| Cell Wall | Cellulose synthase, Pectinase |
| Cell Membrane | Phospholipids, Integral membrane proteins (e.g., G protein-coupled receptors, ion channels), Peripheral membrane proteins (e.g., spectrin, actin) |
| Nuclear Membrane | Nuclear pore proteins (e.g., nucleoporins), Lamins, Nuclear membrane receptors (e.g., importins, exportins) |
| DNA | Histones (H2A, H2B, H3, H4), DNA polymerases, Topoisomerases, DNA repair enzymes (e.g., DNA ligase, DNA polymerase) |
| Cytoplasm | Enzymes (e.g., kinases, phosphatases), Structural proteins (e.g., tubulin, actin), Molecular chaperones (e.g., HSP70, HSP90) |
| Organelle proteins | Proteins present in the Endoplasmic Reticulum, Golgi Apparatus, Mitochondria, Ribosomes and Chloroplast. |
Any of these proteins dissolve the purity of the extracted DNA. For example, the presence of a nuclease degrades the nucleic acid. Any histones left with the DNA decrease the purity of the DNA.
The broad range of Proteinase K activity can remove cell wall, cytoplasmic and DNA-associated proteins. This is important to improve the purity, quantity and integrity of the DNA.
The proteolytic activity of Proteinase K degrades the peptide bonds present in various proteins and makes them inactive. Thereby, it removes contaminants, cell debris and other unwanted cellular components from the DNA.
The graphical representation is given below.
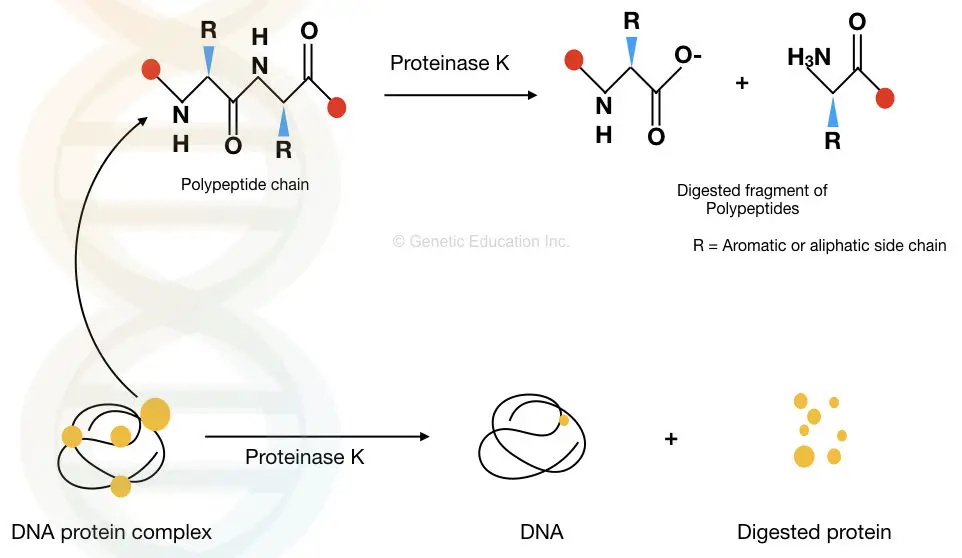
How Proteinase K is used in DNA Extraction?
It’s important to understand that DNA extraction is a complex and multistep process. It uses various chemicals and mechanical methods for effective isolation. Proteinase K alone can not give good-quality DNA.
It is usually added along with the lysis buffer. The cell lysis buffer contains various detergents and chaotropic salts. Common ingredients of DNA extraction lysis buffer are Sodium dodecyl sulphate, EDTA, urea, CTAB, Nonidet P40, NaCl, KCl, etc. Note that detergents play a significant role in DNA extraction.
Moreover, Proteinase K alone can effectively work! Research suggests that it can work greatly with RNase, trypsin and chymotrypsin. In addition, salts such as Sodium Acetate also play an important role in DNA extraction.
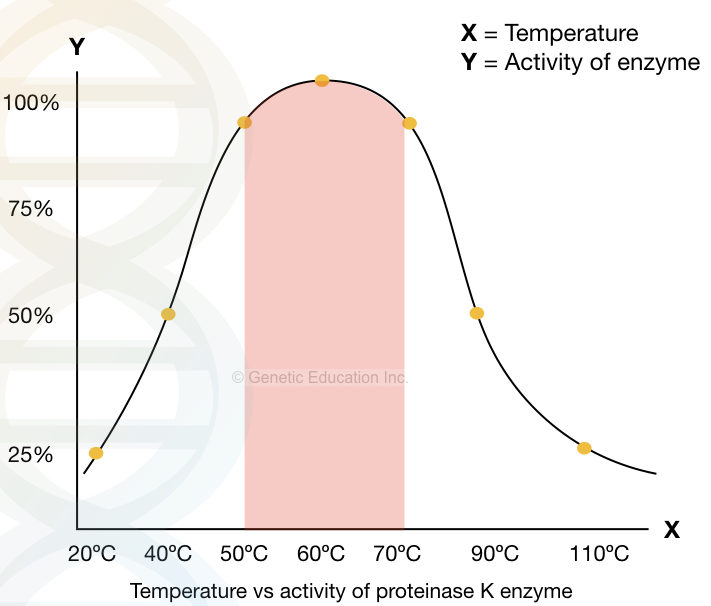
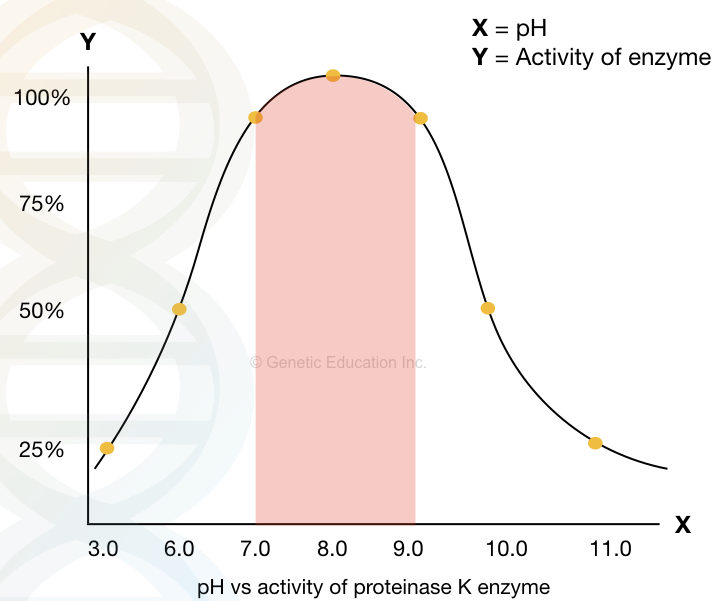
A significant activity of Proteinase K is reported between 37ºC to 70ºC and pH 4.0 to 10.0, however, it works more efficiently at temperatures 60 10 65ºC and pH 8.0. pH 8.0 is the most suitable for DNA extraction.
The graphical illustration of the activity of Proteinase K at various pH.
Concentration
Proteinase K is available in various concentrations. Different experiments need a different concentration. However, the ideal concentration of Proteinase K in DNA extraction is 200μg/ml ranging from 100 to 200μg/ml any concentration we can use, depends on the requirement.
As per our lab experience 200μg/ml gives excellent results. The manufacturer provides Proteinase K in 20mG/ml concentration. Meaning we have to prepare our working solution.
Preparation and Storage
Let’s prepare a working Proteinase K solution by doing some calculations.
20mg = 20,000μg
1ml = 1000μL
20,000μg/ 1000μL is our stock solution (provided by the manufacturer)
The requirement for the working solution is 200μg/ml
C1= 20,000μg/ 1000μL
V1 is a volume given is unknown (?)
C2 is concentration required = 200μg
V2 is volume required= 1000μl
V1C1= V2C2
V1= 1000 * 200/ 20,000
V1= 10
When we add 10μL of proteinase K (from the 20 mg/ml stock) to the 990μL water, the working solution becomes 200μg/ ml, we can use 1μL directly.
Commercially available Proteinase K is provided either in power or in liquid (ready-to-use) form. Prefer to use ready-to-use liquid form, if cost isn’t an issue for you.
If you have a Proteinase K in powder form, you can rehydrate it using either buffer or distilled water. Ideally, a Proteinase K rehydration buffer has been provided with the power but you can use TE buffer, PCR buffer or lysis buffer.
As aforementioned, this stock solution can be further diluted for preparing the working solution.
At room temperature, the enzyme loses its activity over time. Thus the working and the stock solution can be stored at -4ºC and -20ºC, respectively. At -20ºC, it remains inactive for a longer period.
It’s advisable to thaw the solution properly before use. Note that repetitive freezing and thawing may decrease enzyme activity.
Proteinase K DNA Extraction Protocol
Many different variations of Proteinase K DNA extraction protocols exist. Here, I will give you my own standardized and direct-to-use protocol.
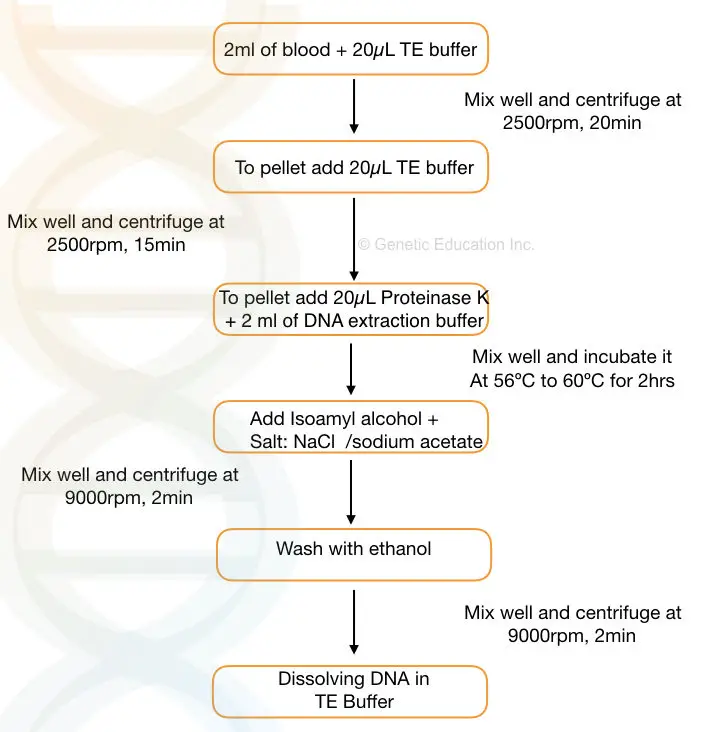
- Take 2 ml of the blood sample and add 10 to 20μL of TE buffer to the sample. Mix well.
- Centrifuge the sample at 2500 rpm for 20 minutes
- Discard the supernatant, add 10 to 15μL of TE buffer to the pallet and mix it gently.
- Centrifuge the sample at 2500 rpm for 15 minutes. Discard the supernatant and add 20μL of proteinase K solution (commercially available ready to use) and 2 ml of DNA extraction buffer to the falcon tube.
- Incubate the sample at 56℃ to 60℃ for 1 to 2 hrs or until the pallet dissolves properly.
- Now add 1 to 2 ml of chilled isopropanol and a pinch of NaCl to the falcon tube and invert the tube for some time to obtain the precipitate.
- Centrifuge the tube at 8000 to 10,000 rpm to settle the pellet at the bottom.
- Discard the supernatant and add 1 ml ethanol and centrifuge it at 10,000rpm for 1 to 2 minutes.
- Discard the ethanol and dry the pellet in the dryer now add TE buffer (pH 8.0) to the pellet and put it in a water bath for 2 to 3 hrs.
This protocol is for 2 ml sample, you can design your protocol for 1 ml or even 500μL sample. calculate each chemical accordingly.
Table: Proteinase K incubation temperature, time and efficiency of digestion
| Temperature (°C) | Incubation Time (minutes) | Digestion Efficiency |
| 37 | 30 | Moderate |
| 37 | 60 | High |
| 50 | 15 | Moderate |
| 50 | 30 | High |
| 55 | 10 | High |
| 55 | 20 | Very High |
| 60 | 5 | High |
| 60 | 10 | Very High |
Advantages
- Widely accepted: Almost all DNA extraction protocols include Proteinase K in their protocol. Research suggests that it is used for various samples including cells, tissues and biological fluids.
- Broad range: Not only in DNA extraction, it’s also used in RNA extraction as well.
- Inactive nucleases: One of the major roles of Proteinase K is the inactivation of nucleases. DNase and RNases are common nucleases that can cleave DNA and RNA, respectively and reduce their integrity.
- Proteinase K can effectively inactivate nucleases and prevent nucleic acid integrity.
- Broad temperature range: Proteinase K can work effectively at higher temperatures as well. This allows DNA extraction under various temperature conditions.
- Minimal inhibition: It is less prone to inhibition by contaminants. Meaning in the presence of inhibitors such as polysaccharides, it can work effectively.
Limitations
- Prone to inactivation: Commonly used chemicals such as phenol or EDTA are potent inhibitors of Proteinase K.
- Moderate specificity: Proteinase K can not completely inactivate all the proteinase. It needs additional detergents and chemicals such as SDS and CTAB for complete lysis.
- Storage: The enzyme can be stored at a lower and freezing temperature. At room temperature, it loses its activity gradually.
- Cost: In addition, the use of proteinase K increases the cost of the experiment.
Optimizations and Modification
In molecular genetics, no single protocol or technique is universally accepted for all the samples and tissue types. Minor to major modifications as per the requirement of the experiment is needed.
The native Proteinase K DNA extraction protocol can be used in combination with other protocols such as Phenol: Chloroform: Isoamyl alcohol DNA extraction protocol. This will increase the accuracy and yield.
For instance, in our protocol, after incubating with the Proteinase K we can use the PCI step. Keep in mind that phenol can reduce the activity of Proteinase K.
It can be used in plant DNA extraction along with the CTAB and RNase. Those who are dealing with plant DNA extraction know how and why it is difficult to extract DNA from plants.
The use of Proteinase K in plant DNA extraction greatly increases the yield and purity.
Lately, chemicals like Triton X 100 and Nonidet P40 can also be used in the Proteinase K DNA extraction protocol.
Advanced Optimizations:
Recently Qiagen has developed a unique formulation for Proteinase K. Now, you can store the liquid and stock Proteinase K solution at room temperature for up to a year.
It comes with the Qiagen spin column DNA extraction kit. The kit gives a good amount of DNA and purity. You can use it.
Reading list:
- DNA Extraction From Dried Blood Spot Samples: Protocol + Comprehensive Guide.
- How To Extract DNA From Whole (Fresh), Frozen, Dried and Clotted Blood?
- How to Extract DNA from Bacteria? + Protocol.
- Comprehensive Guide to Plasmid DNA Isolation.
Wrapping up
I worked on various manual and kit-based DNA extraction methods. Although I personally recommend manual methods, you can use kits which save time for you. Proteinase K is one of my favorite methods.
Trust me with our protocol with proteinase K, you will always get a good amount of DNA with an excellent purity. Give it a try and let me know in the comments. I hope you like this article. Please share it and subscribe to our blog.
Sources:
- Qamar W, Khan MR, Arafah A. Optimization of conditions to extract high quality DNA for PCR analysis from whole blood using SDS-proteinase K method. Saudi J Biol Sci. 2017 Nov;24(7):1465-1469. doi: 10.1016/j.sjbs.2016.09.016. Epub 2016 Sep 10. PMID: 30294214; PMCID: PMC6169501.
- How Proteinase K Can be Used in Blood DNA Extraction by Sigma Aldrich (Merck).
- Gautam, A. (2022). DNA Isolation by Lysozyme and Proteinase K. In: DNA and RNA Isolation Techniques for Non-Experts. Techniques in Life Science and Biomedicine for the Non-Expert. Springer, Cham. https://doi.org/10.1007/978-3-030-94230-4_11.



It is Great blog post. I like it. Thanks for sharing these information with us.