“A DNA sequencing technique is a molecular genetic tool used to determine the sequence of DNA.”
In other words, we can say that the order of nucleotides A, T, G and C in a DNA sequence can be discovered using the DNA sequencing method.
Deoxy sugar, triphosphate and bases are three molecules that construct the DNA in which adenine, guanine, cytosine and guanine are nitrogenous bases.
The polynucleotide DNA chain is made of multiple polynucleotide units thus a nucleotide is a unit of DNA.
Collectively, the sugar, phosphate and base are called a nucleotide which forms a DNA sequence.
Read more: DNA story: The structure and function of DNA.
By determining the order of nucleotides in a DNA sequence we can find out any abnormality related to DNA or gene.
The present article is a very basic article for the novice who actually doesn’t know what DNA sequencing is.
The content of the article is
Key Topics:
Definition of DNA sequencing:
“A DNA sequencing method is used to determine the nucleotides sequence- adenine, guanine, thymine and cytosine, on a DNA sequence orderly, using the fluorescent dye.”
In other words, we can also say, “a nucleotide order of a gene is precisely identified using the DNA sequencing technique.”
What is DNA sequencing?
The nucleotides- A, T, G and C, are arranged in an orderly manner on DNA and it is important for forming a protein.
Hence, the order of nucleotides is very important for studying what protein a gene forms.
PCR only tells us whether a gene or DNA sequence is present or absent, not more than that. On the other hand, A DNA sequencer reads each nucleotide in it.
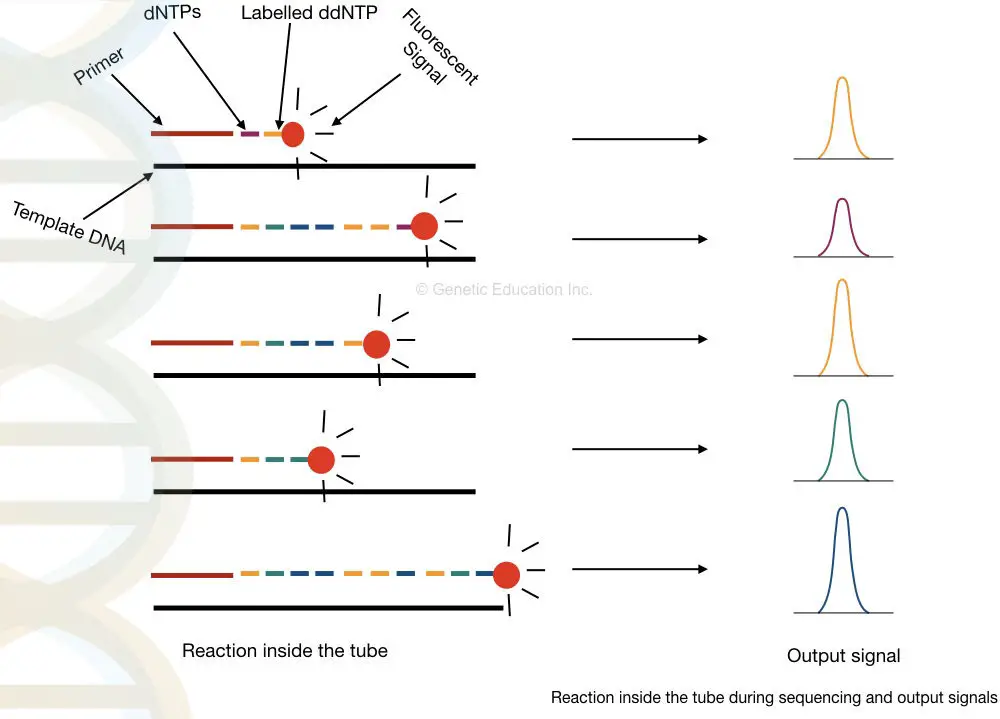
Here, the DNA sequencing method is actually based on the amplification of DNA but instead of the only normal dNTPs, the labeled dNTPs or ddNTPs are used in it.
Once the dNTP binds to its complementary sequence, it emits a fluorescence which is detected by the detector.
Four different fluorescent molecules are used for labeling four different dNTPs thus each labeled dNTP generates different fluorescent signals.
Based on the amount of fluorescence emitted, a peak of it is generated which indicates the presence of a particular dNTP.
Similarly, four different peaks are generated for four different dNTPs. See the image below,
Now, you understand that the labeled nucleotides are one of the ingredients in DNA sequencing. Unlike PCR labeled nucleotides are used in DNA sequencing.
The next reagent used in it is DNA primers. As with the PCR primers, the primers used in DNA sequencing must follow standard primer designing guidelines. Read our PCR primer design guidelines: PCR primer design guidelines.
Depending upon the concentration of template DNA, the concentration of primers varies.
The list of different-sized fragments and their concentration in a reaction is given in the table below,
Table 1:
| Template DNA | Concentration |
| 100-200bp PCR product | 1-3ng/μl |
| 220-500bp PCR product | 5-10ng/μl |
| 501- 1500bp PCR product | 20ng/μl |
| >1500bp PCR product | 30ng/μl |
| <4kb plasmid | 50ng/μl |
| 4kb to 10kb plasmid DNA | 80ng/μl |
| >10kb Plasmid DNA | 100ng/μl |
| Single-stranded DNA | 25-50ng/μl |
| Double-stranded DNA | 150-350ng/μl |
| Bacterial genomic DNA | 2-3μg/μl |
| BAC, cosmid | 0.5-1μg/μl |
The reaction mixture contains all the essential ingredients and enzymes for the amplification during DNA sequencing. Now, for achieving amplification, a Taq DNA polymerase along with the reaction buffer is used in the preparation of the DNA sequencing reaction.
The final makeup of the reaction is done using the dd/w.
This is the general setup of the DNA sequencing reaction. The concentration and volume of each component is given in the table below,
Table 2:
| Component | Concentration | Volume |
| Template DNA | Depends on the assay | 2μl |
| Primers | 10pmol/μl | 2μl |
| dNTPs and ddNTPs mix | 1X | 4μl |
| Buffer | 1X | 4μl |
| Deionized water | 8μl | |
| Total | 20μl |
Alike the PCR cycles, the amplification for DNA sequencing reaction is set up at different temperatures in different steps.
The DNA sequencing cycling conditions,
Table 3:
| Temperature and time | Number of cycles |
| 96°C for 2 minutes | 1 |
| 96°C for 10 sec
50°C for 5 sec 60°C for 4 minutes | 30 |
| 4°C | Hold |
This is the general DNA sequencing setup. Not necessarily applicable for all reactions.
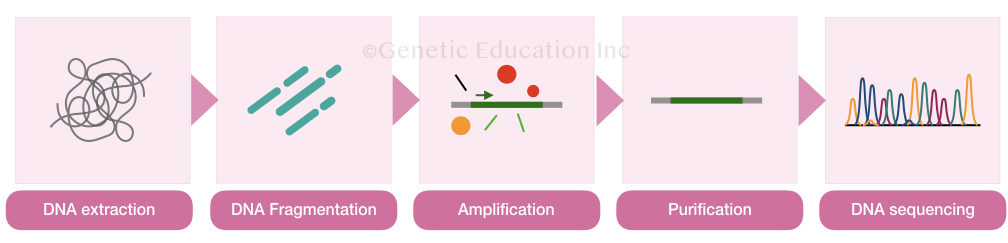
Different steps in DNA sequencing are:
- DNA extraction
- DNA purification
- DNA amplification
- Library preparation
- Sequencing pre-preparation
- Reading the sequencing
- Data analysis
We have already discussed each step in our previous article. Please read the article here: DNA Sequencing: History, Steps, Methods, Applications And Limitations.
An overview of DNA sequencing machine:
The first generation DNA sequencer machines are less machine more manual. Starting from DNA extraction to reaction preparation, all we have to perform manually.
Only amplification and sequencing are done in the machine, after that, the gel must be loaded on a PAGE manually.
Only, 300bp can be sequenced within 12 hours thus the method was tedious and time-consuming.
The next-generation sequencer machines are more advanced, robust and automated. In the present part of the article, I will give you an overview of the sequencer machine, not targeting any specific machine.
Different machines are working on different principles and mechanisms, we are here discussing a general automated sanger-sequencing machine.
The machine has parts or apertures for reaction preparation (buffer system), amplification, capillary electrophoresis and detection. Along with that camera, fluorochrome detector or readers are there in the machine which is attached to the computer.
In addition to this, the machine has a cathode buffer chamber and anode buffer chambers.
The computer has the software for result analysis. This is the simple setup of an automated sanger sequencer.
Read our gigantic article on DNA sequencing: DNA Sequencing: History, Steps, Methods, Applications And Limitations.
How does DNA sequencing work?
I am giving you a brief technical overview so that you can understand what actually we are doing during DNA sequencing.
Before understanding that, keep in mind one thing, different platforms have different working SOPs, also, it may vary from manufacturer to manufacturer.
In addition to this, the manufacturer provides all the reagents and kits for doing DNA sequencing, thus we don’t have an idea about what exactly there in the buffer or mix but I will try to give you a brief outline about it.
The process of sequencing starts with template preparation. The template DNA- BAC, PCR amplicons, plasmid DNA or other DNA fragments are purified first and quantified.
Read our article on PCR template DNA: What are the properties of PCR (template) DNA?
The template quantity chart is given in Table 1 above.
Prepare the reaction mixture as given in table 2 above. Once the reaction is prepared, put it in the thermocycler for amplification as followed by table 3.
Once the amplification is done, now purify the amplicons using any of the given methods by the manufacturer.
I strongly recommended using the ethanol/EDTA precipitation method for the purification of amplicons. amplicon purification is a very crucial step to remove all other unbounded dNTPs, enzymes and other buffer ingredients that might hinder sequencing.
After that, load the sample for capillary electrophoresis.
As the sample migrates, the fluorescent signal generated by it is recorded by the detector which sends it to the computer.
A pre-installed program converts the signal into a “peak”, different peaks for different signals.
Finally, the results can be used further for comparing it with other sequencing or other bioinformatics analysis.
These are the actual technical steps in the DNA sequencing process.
Read further: Differences between Genotyping vs Sequencing.
Examples of DNA sequencing:
16S and 18S rRNA sequencing:
One of the most routine use of DNA sequencing is the identification of pathogenic strains using the 16S and 18S rRNA of pathogens. It is one of the best examples of DNA sequencing used in microbiology- identification of microbes.
The method is rapid, effective and more accurate than the conventional microbiology methods. Related article: 16S rRNA: Gene, Sequencing and Importance.
Gene sequencing:
Genes up to several kilobase pairs can be sequences using the automated sanger sequencing method. Gene sequencing is used to determine any sequence variations within a gene and genotyping.
SNP sequencing:
The SNP-single nucleotide polymorphism is a single base variation that occurs within the population which might be associated with several diseases.
An SNP DNA sequencing allows one to screen an SNP database within an entire genome of an organism.
By identifying different SNPs or groups of SNPs, their association with a particular complex genetic condition can be determined.
High throughput sequencing:
Using rapid and most advanced DNA sequencing methods like next-generation sequencing, an entire genome of an organism can be sequenced within a few hours. This one is one of the best examples of the state-of-the-art sequencing method.
Although, the method is more costly.
Some other examples of DNA sequencing methods are given below,
- Maxum and Gilbert method
- Sanger sequencing
- Whole-genome shotgun sequencing
- Pyrosequencing
- Massively parallel signature sequencing
- Polony sequencing
- Clone by clone sequencing
- 454 pyrosequencing
- Single-molecule real-time sequencing
- SOLiD sequencing
- Sequencing by ligation
- Nanopore DNA sequencing
- Sequencing by synthesis
- Ion Torrent semiconductor sequencing
- DNA nano ball sequencing
- Combinatorial probe anchor synthesis
- Heliscope single-molecule sequencing
- Tunnelling currents DNA sequencing
- Sequencing by hybridization
- Sequencing with mass spectroscopy
- In vitro virus high-throughput sequencing
- Microfluid sanger sequencing
- RNAP sequencing
Related article: 3 Of The Best Genome Sequencing Methods.
Uses of DNA sequencing:
The DNA sequencing technique is used for DNA sequence/ gene or genotype analysis.
It is used in disease diagnosis for a single gene and complex genetic disorders.
It is further applicable in genome-wide association studies through SNP sequencing.
Identification of an organism can be possible from the environmental DNA sample using DNA sequencing.
The pathogen can be identified using the 16S and 18S rRNA gene sequencing.
Besides this, the DNA sequencing method is used in forensic science and plant research as well.
Conclusion:
DNA sequencing is required in every genetic lab because of its wide spectrum applications. Besides this, nowadays, it is most popular in prenatal genetic screening and preimplantation genetic studies.
However, the entire sequencing assay is dependable on the manufacturer, all utilities, kits, reagents and instruments are provided by the manufacturer itself thus additional optimization and experimentation are restricted.
We have to depend on the SOPs and protocol provided by the supplier only.