“Here is the complete breakdown of first, second and third-generation sequencing platforms, techniques, and their advantages and limitations.”
We, now, have almost 47 types of different sequencing methods from various companies. Some are good, some are best and some are worse. Some are high throughput and can sequence many genes, while some are low throughput and can seqence a single fragment.
Each technique has its own importance. Two popular techniques scientists and medical professionals use in 2024 are Sanger Sequencing and Next-generation sequencing.
Sanger can be considered as the first-generation sequencing technique, as it was discovered very early, while NGS platforms can be classified into second and third-generation sequencing techniques depending on their applications.
Students usually get confused between these terms (first, second, third and next-generation sequencing). In this article, I will explain to you sequencing generations, their advantages, limitations and applications.
Stay tuned.
| Sequencing Platform | Generation | Typical Read Length Range |
| Sanger Sequencing | First | Up to 800-1000 base pairs (bp) |
| 454 Pyrosequencing | Second | Up to 700-1000 bp |
| Illumina Sequencing | Second | Up to 150-300 bp (short reads)Up to 250-600 bp (paired-end reads) |
| Ion Torrent Sequencing | Second | Up to 400 bp |
| PacBio Sequencing | Third | Up to 10-30 kilobases (kb) |
| Oxford Nanopore Sequencing | Third | 10 to 100 kilobases (kb) |
Key Topics:
What is first-generation sequencing?
Sequencing techniques developed during the 70s and 80s are known as first-generation sequencing techniques. These techniques were basic, time-consuming and costly. Maxam-Gilbert and Sanger sequencing are the two and only first-generation sequencing platforms.

Sanger Sequencing:
Sanger sequencing was developed by F. Sanger in 1977. As we all know, Sanger sequencing works on the principle of chain termination. When a labeled ddNTP is incorporated into a sequence, it terminates the chain synthesis and provides a sequencing signal.
Each fragment runs on a PAGE gel and is prepared for analysis using autoradiography. However, recently developed capillary electrophoresis is an automation for fragment analysis.
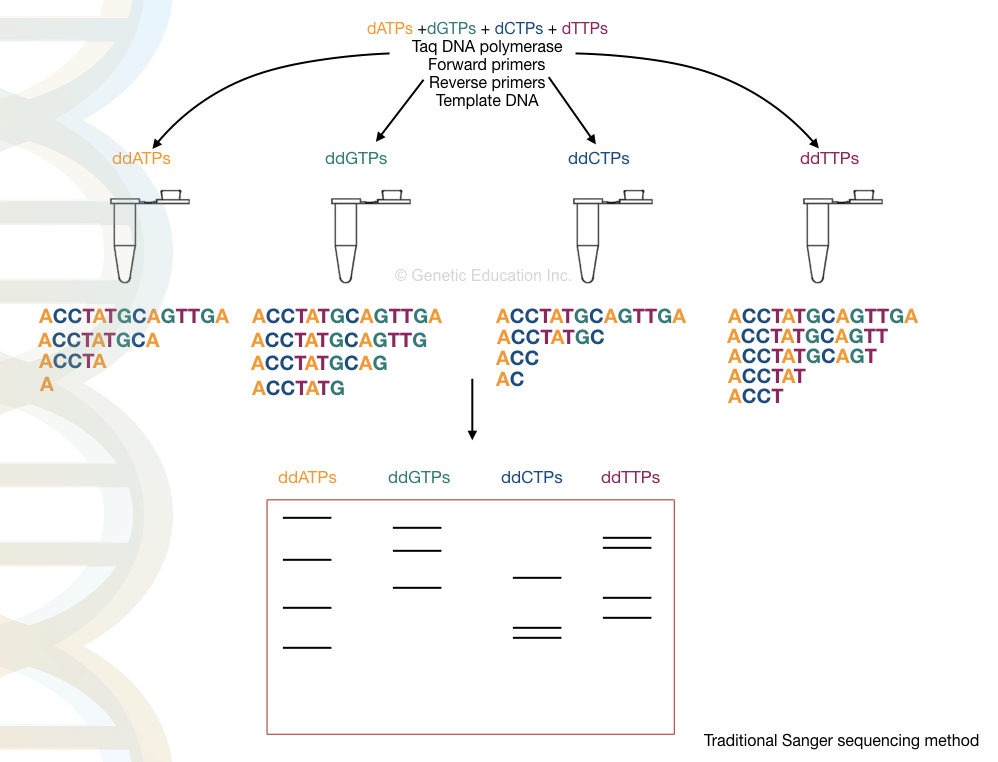
Maxam-Gilbert Sequencing:
The present FGS method also requires gel electrophoresis for analysis. It was developed earlier by A Maxam and W Gilbert in 1970. In this method, the DNA sample is selectively cleaved and treated with chemical reagents.
The chemical reagents modify each nucleotide position, which is once again cleaved and run on a gel for analysis. The present method is a labor-intensive and complicated sequencing method.
Advantages:
Let’s first discuss some benefits.
FGS, for instance, Sanger sequencing is highly accurate. It has an error rate of 1 in 1000 bp. Such accuracy is highly recommended for single gene sequencing or sequencing smaller fragments.
The typical read length of FGS is around 800 to 1000 bp. This benefit also provides flexibility in gene sequencing.
Although Maxam and Gilbert’s method was extinct over time, Sanger sequencing is still used in genetic research and diagnosis. It’s considered as a gold-standard method in diagnosis.
Limitations:
As these are the initial sequencing efforts, these techniques have so many limitations.
In FGS, the sequencing process is followed by gel electrophoresis. That means post-processing is required to study and interpret the results. Scientists, literally, run each sequenced fragment on a gel, take a picture and count and sequence each nucleotide.
FGS has limited sequencing capacity and can sequence up to 1000 or 1200 nucleotides, effectively. For instance, in Sanger sequencing, a few nucleotides from the beginning and end can not be considered in the final sequencing results due to their low-quality reads.
The sequential process of FGS can sequence a single gene or fragment in a single run. That means, are low throughput and can not be used for high throughput analysis.
As aforementioned, FGS is labor intensive, every step is performed manually including post-sequencing gel electrophoresis. However, right now, we have the most automated version of Sanger sequencing.
FGS techniques require hazardous chemicals and reagents. For instance radiolabel dye in Sanger sequencing, and piperidine and hydrazine in Maxam-Gilbert sequencing.
Moreover, FGS sequencing techniques like Sanger sequencing face problems in sequencing homopolymeric sequences. This restricts its sequencing power to only a limited set of sequences.
| Advantages | Limitations |
| High accuracy | Low throughput |
| Long sequencing reads | Labor-intensive |
| Well-established methodology | Limited application for large-scale projects |
| Cost-effective for short reads | Difficulty sequencing repetitive regions |
Related article: What is Long-Read Sequencing?
What is Second Generation Sequencing?
After the ‘90s, the era of new-age and high-throughput sequencing began with the introduction of pyrosequencing. SGS is short-read sequencing, accurate and faster than the FGS.
454 pyrosequencing, SOLiD, Illumina and Ion torrent sequencing are the platforms that evolved as SGS methods. Notedly, SGS methods work on sequencing-by-synthesis chemistry.
| Instrument | Company | Chemistry | Output (GB) | Sequencing Time (Hrs) | Detection |
| iSeq | Illumina | SBS | 0.3-1.2 | 9-17.5 | Fluorescence |
| MiniSeq | Illumina | SBS | 1.7-7.5 | 4-24 | Fluorescence |
| MiSeq | Illumina | SBS | 0.3-15 | 4-55 | Fluorescence |
| NextSeq | Illumina | SBS | 10-120 | 12-30 | Fluorescence |
| HiSeq | Illumina | SBS | 10-1000 | <3 days | Fluorescence |
| NovaSeq | Illumina | SBS | 2000-6000 | 16-44 | Fluorescence |
| PGM | Thermo Fisher | SBS | 0.08-2 | 3-10 | PSC |
| S5 | Thermo Fisher | SBS | 0.6-15 | <19 | PSC |
| Proton | Thermo Fisher | SBS | 10-15 | 4-24 | PSC |
454 pyrosequencing:
Life Sciences developed the pyrosequencing technique and was the first NGS platform with high throughput capabilities. Later on, it was acquired by Roche and named 454 pyrosequencing.
It follows multi-enzymatic steps for producing sequencing reads. Synthesis catalyzed by polymerase releases a pyrophosphate. The PPi is converted into a light molecule in a multi-catalytic process and recorded as a positive amplification signal.
Each time only a single nucleotide is sprayed on a Picotiter plate and recorded on a picogram. The use of multiple enzymes, poor capacity to sequence homopolymers and extensive sequencing are its limitations.
Although it has a longer read length compared to other sequencing platforms (Illumina and Ion torrent).
Due to poor sequencing capabilities and the evolution of higher throughput techniques, 454 pyrosequencing has become outdated.
SOLiD sequencing:
SOLiD sequencing is Sequencing by Oligonucleotide Ligation and Detection. It was developed by Applied Biosystems, later on, acquired by Life Technologies and then Thermo Fisher Scientific.
The chemistry is simple. Labeled oligonucleotides are ligated to the DNA sequence and detected as a positive sequencing signal by the detector. It has a very short read and can be used for SNP detection and resequencing.
Unfortunately, SOLiD sequencing couldn’t stand in the market and was discontinued in 2016. It failed to compete with other high throughput and third-generation sequencing options.
Illumina sequencing:
Illumina is the most powerful player in second-generation sequencing. It offers short-read sequencing with the most accurate fluorescent-based sequencing-by-synthesis chemistry.
iSeq, MiniSeq, MiSeq, HighSeq, NovaSeq and NextSeq are the Illumina NGS platforms that provide output data starting from 1Gb to 6000Gb in a day. The early bird iSeq yields 1.2Gb output data whereas the NovaSeq yields up to 6000Gb/day in up to 44 hours.
One of the major advantages of the Illumina sequencing platform which the previous generation platforms failed to deliver is effectively sequencing the homopolymer block. The SBS chemistry here is designed in such a way that it can only incorporate a single nucleotide at a time.
Even though there are multiple single nucleotides present (for example, AAAA), the complementary nucleotide is added and blocks the incorporation of the next nucleotide until the fluorescence signal is generated, recorded and read.
Illumina sequencing techniques are capable enough to sequence the entire genome in a single day and can identify variants (SNPs, insertions and deletions).
Nonetheless, it requires a high-end and sophisticated computational setup, experts for bioinformatics analysis, and storage and processing capacity to handle such a high amount of data.
Illumination sequencing is high throughput, rapid, cost-effective, labor-extensive and semi-automated.
Ion Torrent:
Ion Torrent was developed by Ion Torrent Systems and later on, acquired by Thermo Fisher Scientific. It works on the principle of SBS but uses semiconductor chemistry. It’s similar to pyrosequencing but detects hydrogen ions instead of pyrophosphate.
During the nucleotide incorporation, released hydrogen ions cause a change in the pH. The semiconductor detector detects the pH change and records the results. It doesn’t rely on fluorescent labeling for detection.
However, it faces the same problem as pyrosequencing. It can not effectively sequence the homopolymer sequence block. However, it’s still accurate, has high throughput and is cost-effective compared to pyrosequencing.
Thus, it is used in amplicon, targeted and microbial sequencing. Ion Torrent can yield 2 to 15 GB data within 24 hours which is pretty lower than the Illumina platforms. However, it has ready-to-use assays for reproductive biology, prenatal testing, and postnatal analysis of various disease and cancer panels.
In addition, Genexus, like Ion torrent platforms, is completely automated. I can extract DNA, prepare libraries, run sequencing, analyze data and generate a report.
Advantages:
SGS has high throughput and can sequence millions of DNA fragments in a single run using massively parallel sequencing.
It’s the most accurate sequencing assay as it works on short-read and paired-end sequencing technologies.
SGS is cost-effective. The per base sequencing cost of the SGS is too low compared with the FGS.
It has a rapid turn-around time. The majority of platforms can sequence the entire human genome within a day. That eventually decreases TAT for diagnostics.
The initial sample requirement is also very low in the case of SGS.
Limitations:
The shorter read length is the strength and the weakness of the present sequencing generation. A shorter read length complicates the genome assembly process and increases the likelihood of errors.
As it works on massively parallel sequencing, there are higher chances of errors during sequencing.
SGS relies on PCR amplification that can introduce bias and artifacts and mislead the sample representation.
These techniques have higher error rates, meaning they can introduce more sequencing errors than Sanger sequencing. However, bioinformatics tools are now available to solve errors.
As aforementioned, second-generation sequencing requires sophisticated bioinformatics setup and computational and data processing resources. Furthermore, it needs high-end expertise and experience to perform analysis.
Lastly, the testing cost with SGS is cheaper but the instrumental setup is expensive. It’s difficult for smaller laboratories to acquire and run such expensive techniques.
| Advantages | Limitations |
| High throughput | Shorter read lengths |
| Cost-effectiveness | PCR amplification bias |
| Rapid turnaround time | Bioinformatics complexity |
| Versatility | Instrumentation and infrastructure cost |
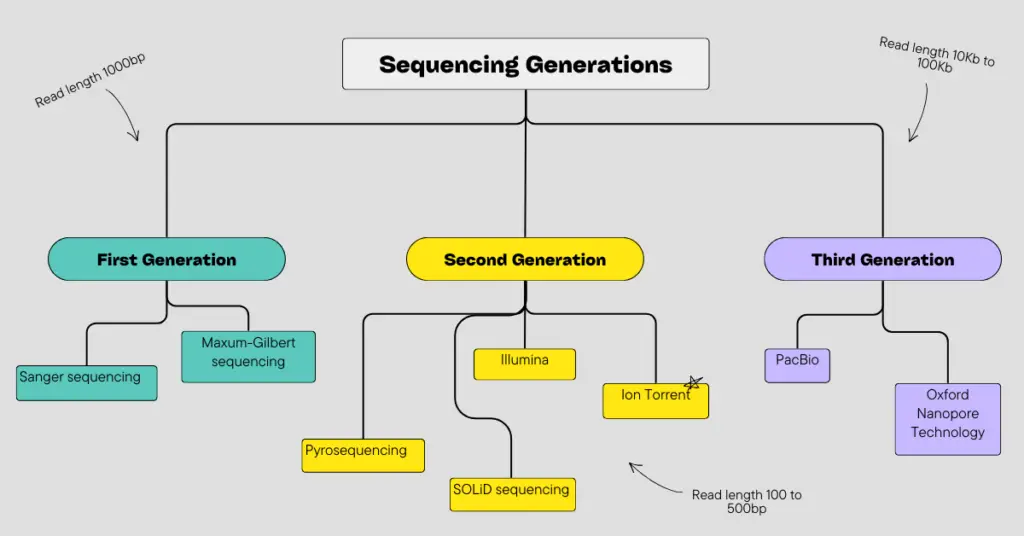
What is third-generation sequencing?
Third-generation sequencing is an ultra-high throughput and long-read sequencing platform that can perform sequencing on a single molecule. It can sequence longer fragments effectively.
The typical read length ranges between 10Kb to >100Kb. It works on the principle of SMRT (Single-Molecule Real-time sequencing), where we can observe sequencing in real-time. The outstanding advantage of TGS is that it can detect epigenetic alterations, like DNA methylation in real time.
TGS is crucial in the coming time as it has the potential to effectively sequence highly polymorphic, repetitive and GC-rich regions. This becomes increasingly important because the major genomic portion is made up of repetitive regions.
Common platforms that provide TGS long-read sequencing are PacBio and ONS (Oxford Nanopore Sequencing).
| Instrument | Company | Chemistry | Output (GB) | Sequencing Time (Hrs) | Detection |
| MinION | ONT | SMRT | 10-20 | 30 min-48 | ICC |
| GridION | ONT | SMRT | 50-100 | 30 min-48 | ICC |
| PromethION | ONT | SMRT | 480-960 | 30 min-48 | ICC |
| RSII | PacBio | SMRT | 0.5-1 | <6 | Fluorescence |
| Sequel | PacBio | SMRT | 5-10 | <20 | Fluorescence |
PacBio sequencing:
Pacific Bioscience offers long-read sequencing using SMTR technology. Here, each incorporated nucleotide is read when it passes through wells having immobilized polymerase.
The fluorescence generated from the synthesis process can be detected in real time. It can sequence a single DNA molecule at a time from a single well. The plate contains thousands of microwells. So each fragment is sequenced, in real-time.
Sequel and Sequel II are two PacBio long-read sequencers.
Nanopore sequencing:
ONT (Oxford Nanopore Technology) uses SMRT along with a change in electrical current for detection. Here, when the DNA is passed through the immobilized protein nanopores (embedded in the membrane), it produces a change in the electrical current.
Each nucleotide incorporation produces a different type of electrical current change, the signal recorder converts electrical current into nucleotides. Nanopore sequencing typically has a 10Kb to 100Kb read length.
Again, each fragment is sequenced in individual protein nanopores, read and converted in real time. MinION, GridION and PromethION are three ONT sequencers.
Advantages:
The longer read length provides better sequencing of complex genomic regions. Although it is difficult to sequence such large fragments, the unique SMTR chemistry allows efficient sequencing.
The unique SMTR concept allows the sequencing of a single molecule at a time which gives better sequencing resolution.
TGS does not need PCR amplification and thus, is resistant to PCR bias and errors.
Real-time sequencing monitoring allows rapid data generation. Such chemistry also supports studying DNA methylation like epigenetic alterations directly.
It effectively studies complex, repetitive, homopolymeric and GC-rich genomic regions.
TGS typically requires fewer sample preparation steps.
It can perform sequencing in rapid time. It can generate <1000bp data within 44 hours.
Limitations:
Longer reads mean higher error rates. TGS are prone to errors.
The instrumental setup and additional requirements increase the overall cost.
The complex set of data generated from the TGS required high-end and sophisticated bioinformatics and computational resources.
TGS is still in the development phase, its full potential has yet to be explored.
| Advantages | Limitations |
| Long read lengths | Higher error rates |
| Single-molecule sequencing | Data analysis complexity |
| Real-time sequencing | Instrument cost |
| Reduced sample preparation | Technological immaturity |
Related article: 10 Challenges in Whole Genome Sequencing.
Conclusion
In conclusion, each generation of sequencing platforms has its advantages and limitations. However, second and third-generation sequencing, collectively known as NGS, is more advantageous.
In recent times, NGS allowed us to sequence the entire genome in a day. Such a huge sequencing breakthrough opened new doors for pharmacogenomics, nutrigenomics and transcriptomics studies.
I hope you like this article. Do share it and subscribe to our blog.
Sources:
Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics. 2016 Jan;107(1):1-8. doi: 10.1016/j.ygeno.2015.11.003. Epub 2015 Nov 10. PMID: 26554401; PMCID: PMC4727787.


