“Genome sequencing is a state-of-art, robust and high throughput technique to sequence the entire genome of an organism. It enables scientists to study the entire genetic composition of an organism. Let’s find out what genome sequencing is and 3 common methods.”
Snippet:
- The Human genome contains ~3.2 billion base pairs, ~30,000 genes and 97% non-coding sequences.
- The entire genome is arranged on 23 pairs of chromosomes.
- Only ~3% of the human genome can code for proteins, the rest are just junk.
Genome sequencing became popular in the recent COVID era. Instead of sequencing a viral gene, sequencing the entire tiny genome of a virus provides pretty much more information. However, until now, scientists are using it to study the genome of humans, animals or plants.
Such organisms have a huge genome, thousands of genes and enormous junk DNA. It makes the task tedious, time-consuming and costly. Hence wasn’t penetrated into mainstream science and remained limited to genetics research only.
Now scientists and medical professionals can use it for diagnosis, study pandemics and epidemics, and to understand how different strains evolve and develop resistance. However, genome sequencing in recent times is superior to previous assays.
Sequencing a genome of SARs-CoV2, H1N1 influenza or HBV improves our understanding of the dynamic nature of the pathogens, their pathogenicity and how they evolved. So it is important to understand what genome sequencing is, how it is different from gene sequencing and what are the best 3 methods.
I’m Dr. Tushar, a scientist, a scientific writer and an expert in genetics and genomic technologies. I will answer all of these questions in this article.
Stay tuned.
Related article: 10 Challenges in Whole Genome Sequencing.
Key Topics:
What is genome sequencing?
Genome is like a whole book; a huge, gigantic repository in terms of volume and information as well. So a codon is a word, a sequence is a sentence and a gene is a page from a book.
What gives more information? A book, right? As we read each letter of the book, we acquire more and more information. On the other hand, reading a page or sentence has limited or partial information.
Henceforth, reading a book will be more beneficial and the same is true for the genome too. Genome has huge information, however, decoding genomes like humans, plants or other animals is a tedious, time-consuming and laborious process.
So technically, sequencing, or reading an entire genome rather than only a gene or some unknown sequence, is referred to as genome sequencing. Now first see the differences between DNA/gene sequencing vs genome sequencing.
Gene vs genome sequencing:
| Gene sequencing | Genome sequencing |
| Sequence a gene of only some thousand base pairs. | Sequence the entire genome of an organism. |
| Provides information of a protein-coding gene and related alterations. | Provides information regarding different genes, non-coding regions, introns, exons, transposons and other genomic components. |
| Technically a more straightforward process includes DNA extraction, amplification, sequencing and reading. | Technically more complex processes including DNA extraction, amplification, library preparation, adapter ligation, sequencing, conting and reading. |
| Handy and cost-effective process. | Complex, costly, laborious and tedious process. |
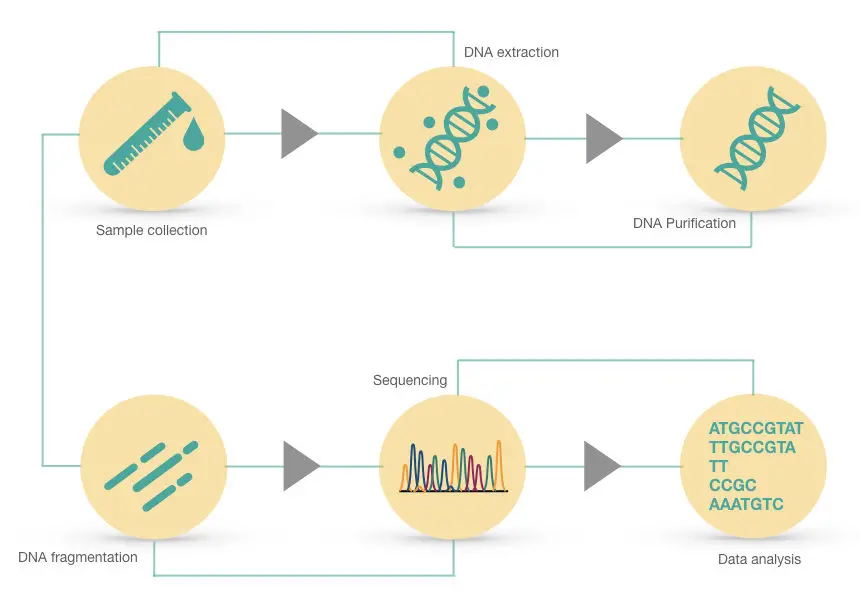
Steps and process of genome sequencing:
The process of genome sequencing is a bit more complex than gene sequencing, as we talked and thus required an expert hand. The process completes in the following steps.
DNA extraction, DNA fragmentation, library preparation, amplification, sequencing, reading and analyzing results.
Fun fact:
If you print all of the nucleotides of the human genome it would fill 4200 books.
DNA extraction: The extraction process gives us pure and high-quality DNA which is an utmost requirement for any sequencing assay. Notedly, genome sequencing needs even a higher quantity of DNA.
If you wish to learn more about DNA extraction, various techniques, methods and protocols please visit this category from our blog: DNA extraction.
DNA fragmentation: Now this is important to understand. We can’t sequence the entire genome in a single run. It’s huge, I mean very huge. Further to this, a machine also has a limited detection range. So overall, understand that reading the entire genome isn’t possible.
What scientists do is that they perform fragmentation and make different known-sized fragments of the genome. Now each fragment is read or sequenced simultaneously. Usually, restriction digestion-like techniques are used for this.
Library Preparation: Genomic library preparation is an important part of this process and thus we have written a dedicated article on this. You can read it by clicking the link. By definition, the genomic library is a collection of same-sized/ known DNA fragments.
Each end of fragments is ligated with adaptors which are known sequences, complementary to the sequencing primers.
- Related article: DNA fragmentation- Techniques, Importance and Applications.
Amplification: In the next steps, each fragment is amplified in a PCR to generate a sufficient amount of copies for sequencing. Note that amplification is often known as library enrichment.
- Related article: 10 Strategies to Achieve Excellent PCR Amplification.
Sequencing: Now the machine uses the algorithm installed, and each fragment from the amplified library is sequenced, meaning read by the machine. The detector detects signals for each nucleotide and collects the sequence information.
Reading, conting and interpretation: Now the sequence data of all fragments are gathered, re-joined by contings and sent for bioinformatic analysis. Our entire genomic data is now ready for analysis.
3-Best genome sequencing methods:
Next-generation sequencing:
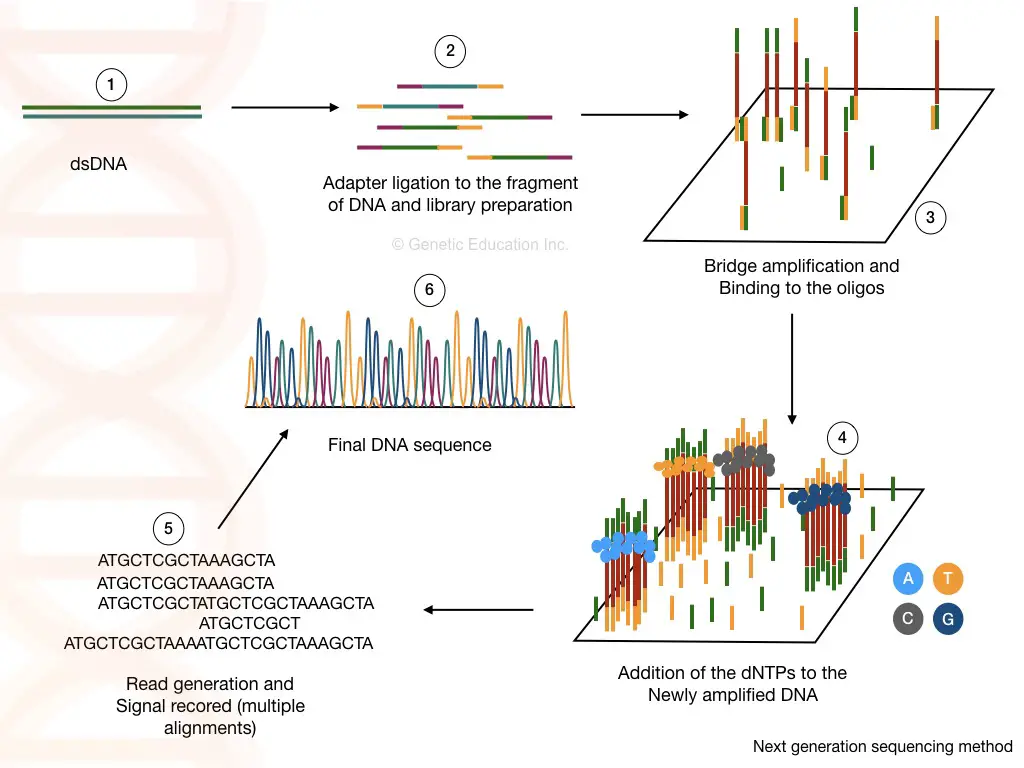
In recent times, next-generation sequencing, abbreviated as NGS, is the most advanced, robust, accurate, faster, cheaper (comparatively) and high throughput genome sequencing technique. It relies on the chemistry of bridge amplification.
Note:
NGS can sequence more than 5 separate human genomes simultaneously in a single run at a cost of around $5,000.
The sequencing occurred in a massively parallel amplification of fragments through bridge amplification. Although, the overall success of the NGS highly relies on the genomic DNA library preparation.
As aforesaid, the genomic library is the collection of genomic DNA fragments of the desired length. Adapter ligation and barcoding for identification are crucial steps in this process. Multiplex-sequencing occurs by pooling different libraries in a single run which saves time and resources.
A specialized library-specific barcode sequence helps to distinguish libraries of different samples, after sequencing. Using sequencing by synthesis phenomenon, each nucleotide is read in a flow cell.
A detector detects the fluorescence signal for each nucleotide and arranges it accordingly. This process is technically denoted as bridge amplification.
“The NGS is the most trusted genome sequencing method so far and trusted by experts. The bridge amplification process by “sequencing by synthesis” makes it unique, reliable and most accurate”, said Dr. Jigar Suthar from the Unipath Laboratory, Ahmedabad, India.
Clone by clone sequencing
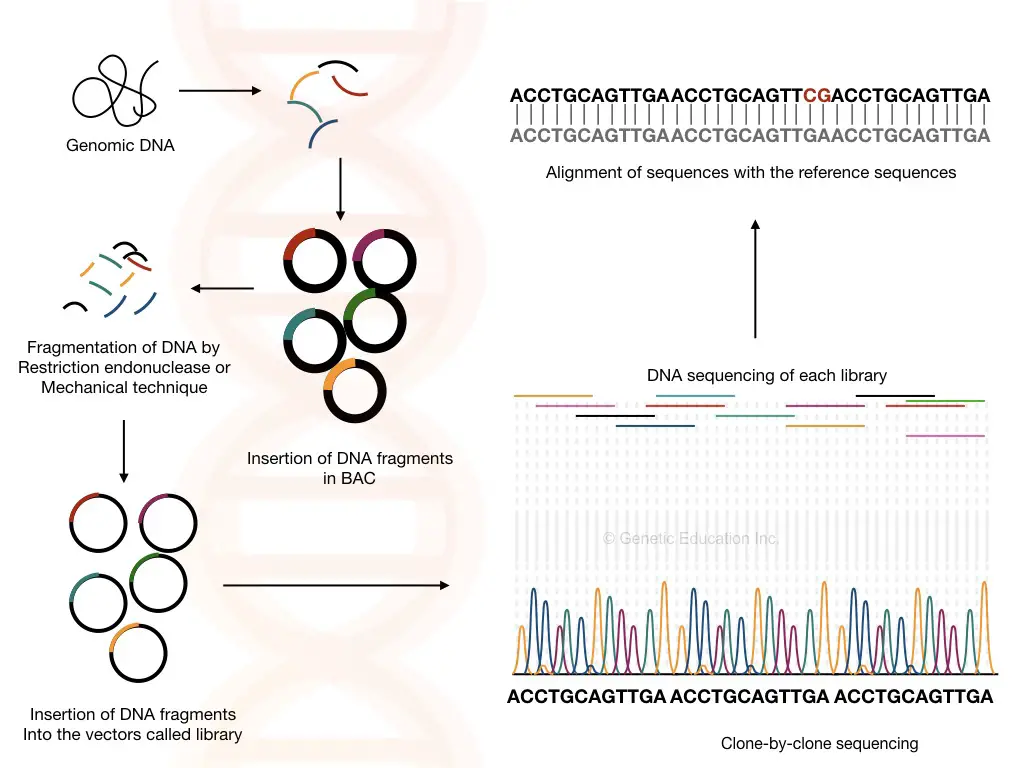
The clone by clone sequencing method for genome sequencing is a traditional technique, used during the 90s to sequence smaller genomes like C. elegance, however, it was also used during the human genome project too.
Sequences are first mapped on a chromosome to know the exact location and fragmented. Larger fragments of 1500bps are now ready to insert into a BAC vector. In bacterial artificial chromosomes, it replicates and generates millions of copies.
The fragments are further clumped into a few hundred base pairs and allowed to insert into a known plasmid followed by sequencing.
The overlapping known plasmid or vector sequences are removed, and the genome sequences are collected and mapped back on the chromosome using the prior mapping data.
The clone-by-clone sequencing is traditional, laborious and time-consuming, nonetheless, it is more accurate. Interestingly, for larger genomes like eukaryotes, it works excellently. The requirement of prior location data or chromosome map is a major limitation here.
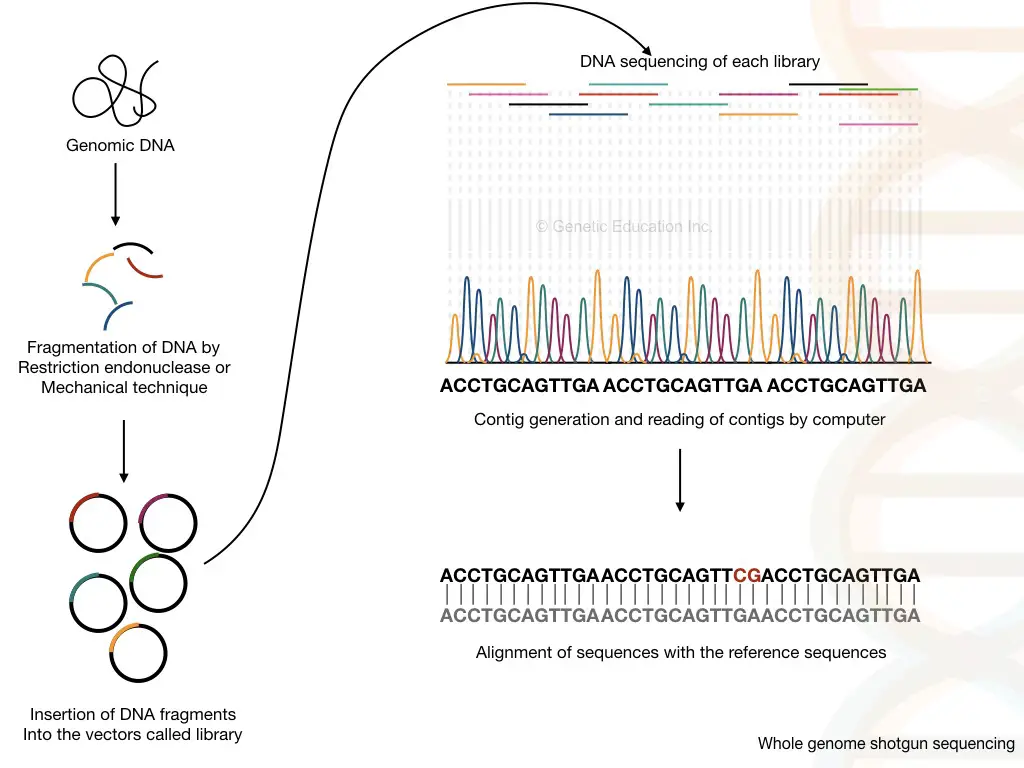
Whole genome shotgun sequencing
Technically, I can say, the shotgun sequencing method is the improved version of clone-by-clone sequencing which is strength forward and speedy and doesn’t require cloning and mapping.
The entire eukaryotic genome is broken down into fragments of 2000 to 20,000 base pairs, incorporated in a library and sequenced by amplification. Fluorescently labeled nucleotides when incorporated terminate the chain reaction and emit fluorescence.
The detector detects the signal and arranges the sequence, accordingly. In the final step, the computer program arranges the sequence of all the fragments and generates the whole genome sequence.
Originally, the method was conceptualized and used by Sanger et al. to sequence the whole human genome. Notedly, it works on the principle of chain termination.
Cost of whole genome sequencing:
As per the information available on the internet, the entire cost of the human genome project was ~3 billion dollars. But let me tell you that you do not have to pay such a huge amount to sequence your genome.
The genome sequencing cost varies from $500 to $5000 per sample depending upon the length of the genome, the technology selected and the method used. For example, The NGS is comparatively costlier than other methods.
In the US, it costs around ~$300 to $1000 per sample. While in India it cost around 20,000 to 5000 Rs per sample.
Wrapping up:
These three techniques gained honor in sequencing technology as were used during the human genome project. Nonetheless, each has its own advantages and limitations. So which technique to use highly depends on the experience of the researcher.
I tried to explain things in layman and focused on only technical aspects so that students can understand how things work in the lab. If you want to learn more on sequencing, you can read our previous article. This article contains almost all the information regarding every possible sequencing technology.
Related article: DNA Sequencing: History, Steps, Methods, Applications and Limitations.