“Curious about what causes genetic disorders? Decoding 15 common mechanisms causing genetic disorders.”
Genetic disorders are caused by abnormalities in our genetics either in the DNA, RNA or chromosome. It can result in serious, congenital and inherited— mental, psychological, developmental, neurological and motor disability consequences.
As per the WHO report, the global burden of genetic disorders is 2 to 5% (new births/ every year). However, the National Human Genome Research Institute reported that ~350 million people are living with rare genetic disorders on Earth (1, 2).
More to this, the frequency of various birth defects and congenital conditions is increasing every year. So genetic disorders are a serious health concern for us. Along with genetic factors, other factors and mechanisms contribute to this condition.
What are those?
In this article, I will explain 10 mechanisms (with examples) that result in various types of genetic disorders. Keep in note that we are not explaining what genetic disorders are! we will directly explain the causes.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What Causes Genetic Disorders?
Any genetic disorder is caused by alterations in either DNA, chromosome, or gene expression. Mutation or alteration refers to any change in the structure of a DNA or chromosome.
Common alterations are gene mutations, SNPs, chromosomal alterations (structural or numerical), CNVs, repeat expansion, mitochondrial mutations, etc.
Other factors, that contribute to causing genetic disorders are gene-gene interactions, genetic instability, genomic imprinting, repeat expansion, transposon activities, non-coding RNA dysregulation, copy-neutral loss of heterozygosity, errors during DNA metabolism and gene-environment interaction.
We will elaboratively explain every mechanism listed here with an example.
Gene Mutations:
Gene mutations are alterations in the DNA sequence or gene that can occur spontaneously or due to external factors. Among many possible gene mutations, insertion, deletion, duplication or translocations are so common.
Other types of mutations can be categorized as missense, nonsense, frameshift, and silent mutations, each with different consequences on the final gene product. For example, a single base change in a gene may result in a genetic abnormality
CFTR gene mutation associated with cystic fibrosis is one of the well-known examples of gene mutations. In individuals with cystic fibrosis, mutations delta f508 lead to the production of a defective or non-functional protein.
This mutation disrupts the normal function of the CFTR protein. The consequences of the present condition affect the transport of chloride ions across cell membranes and resulting in the buildup of thick, sticky mucus in the respiratory and digestive systems (3).
Related article: Genetic Mutations- Definition, Types, Causes and Examples.
Single Nucleotide Polymorphisms (SNPs):
Alterations at a single nucleotide position, either insertion or deletion in the DNA sequence are known as Single Nucleotide Polymorphisms (SNPs). SNPs are usually non-pathogenic and can be repaired by our own DNA repair mechanism but can cause several serious consequences in some conditions.
SNPs are the most common type of genetic variation that can produce differences in traits, susceptibility to diseases, or response to medications. However, to qualify as an SNP, it should typically present in at least 1% of the population.
The FTO gene is associated with obesity. rs9939609 SNP in this gene causes varied degrees of obesity. It is a T>A mutation in which individuals carrying the A allele of this SNP may have a higher risk of obesity compared to those with the T allele.
Studies also revealed that the present SNP is also involved in regulating appetite and energy expenditure (4).
Chromosomal Alterations:
Chromosomal alterations are changes in the structure or number of chromosomes. This can include deletions, duplications, inversions, or translocations, and are larger than gene mutations.
These alterations can result from errors during cell division or exposure to environmental factors. Trisomy 21, also known as Down syndrome is a well-known chromosomal abnormality that occurs by a change in chromosome number.
In individuals with Down syndrome, there is an extra copy of chromosome 21, along with the pair. Down syndrome leads to developmental and intellectual disabilities, as well as characteristic physical features (5).
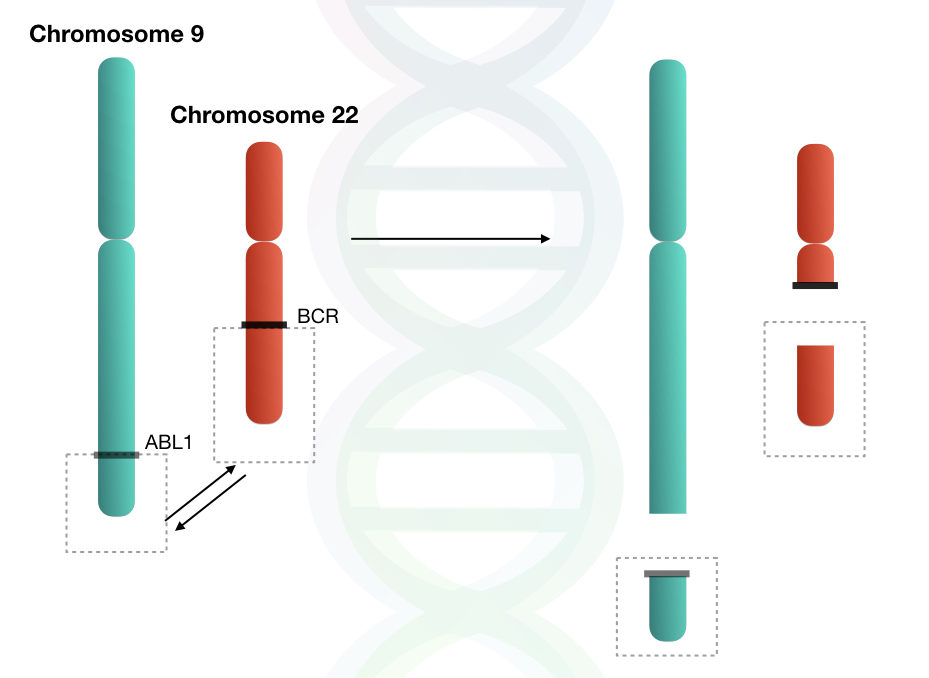
Philadelphia chromosome, involving translocation of genetic material between chromosomes 9 and 22 is an example of a structural chromosomal alteration (6).
Copy Number Variations (CNVs):
Variations in the number of copies of a particular DNA or gene segment are known as CNVs or Copy Number Variations. CNVs can involve deletions or duplications of relatively large genomic regions and can have significant effects on gene dosage.
For example the 16p11.2 region. This particular region has been associated with a range of neurodevelopmental disorders, including autism spectrum disorder (ASD). Copy number variation in this region causes changes in the dosage of multiple genes, influencing brain development and function (7).
Gene-Gene Interactions:
A gene’s normal function is to produce a protein. For a pathway, many genes contribute their sole purpose. However, oftentimes, interaction between genes causes serious complications.
The phenomenon is known as epistasis in which the gene-gene interaction masks or modifies the effect of another gene. Now this can influence the overall phenotype and the risk of developing certain traits or disorders.
One such example of gene-gene interaction and its negative consequences is an interaction between the APOE and the PSEN1 gene in Alzheimer’s disease. The ε4 variant of the APOE gene is a known Alzheimer’s risk variant.
The age of onset and disease severity depend on the interaction of the APOE variant and PSEN1 gene (8).
Epigenetic Modifications:
Epigenetic modifications are changes in gene expression and not in gene structure. How many times does a gene express or produce a protein is referred to as gene expression.
DNA methylation, acetylation, phosphorylation, histone modification and chromatin remodeling are some common epigenetic modifications or tags. If any epigenetic tag is modified, for example, methylation pattern, it results in abnormal gene expression.
Alterations in gene expression for a gene or group of genes cause serious health-related problems. Cancer is the most common example of epigenetic modification. Hypermethylation of certain tumor suppressor genes can lead to gene silencing, resulting in uncontrolled cell growth and cancer (9).
Related article: Epigenetics 101: What is Epigenetics and How Does It Work?
Gene-Environment Interactions:
Gene-environment interaction is the most common cause of genetic disorders. Even if it is a gene mutation or a chromosomal alteration, it occurs due to an adverse environment. For example, exposure to toxins, radiation, etc.
Gene-environment interactions refer to the interplay between genetic and environmental factors. Such interactions determine traits, phenotypes, or susceptibility to diseases. Diet and lifestyle are other known environmental factors that influence genetics.
Adding to our previous example of the APOE gene, the interaction between the APOE gene and dietary fat intake influences the cardiovascular health of an individual. Here the APOE gene is a genetic factor while the dietary fat intake is an environmental factor, collectively causing serious cardiovascular problems (10).
Related article: Influence of Gene-Environment Interaction on Life.
Genetic Instability:
Genetic or genomic instability is a condition of high-frequency mutations within the genome such as gene mutations, chromosomal rearrangements, or aneuploidy. Meaning, an individual’s genome becomes more susceptible to acquiring various mutations.
Although many factors contribute to genomic instability, defective DNA repair mechanisms and mutagen exposure are the two most common factors. For example— Lynch Syndrome.
Lynch syndrome (also known as Hereditary nonpolyposis colorectal cancer) has an increased risk of developing colorectal cancer due to mutations in DNA mismatch repair genes (11).
Related article: Role of Microsatellite Instability (MSI) in cancer (MSI-H, MSI-I and MSS)
Genomic Imprinting:
I defined the term genomic imprinting as the expression of a gene or both alleles from only a single parent, either from a father or mother. So for an imprinted gene, only a single parent contributes the alleles; this leads to serious genetic abnormality.
Genomic imprinting is an epigenetic mechanism that results in the selective silencing or activation of specific alleles based on parental origin. To know more, you can read our series of articles on this topic.
Topic: Genomic Imprinting.
Two classic examples of genomic imprinting are Prader-Willi syndrome and Angelman syndrome. These two conditions are the first case of genomic imprinting due to mutation in a specific region of chromosome 15.
On chromosome 15, Imprinting of the maternal side results in Angelman syndrome while imprinting from the paternal side results in PWS (12).
Repeat Expansion:
Another interesting mechanism that leads to genetic abnormalities is repeat expansion. Our genome is highly repetitive and consists of various repeating units. Repeat expansion refers to the abnormal increase in the number of DNA repeat sequences within a gene.
These expanded repeats can interfere with normal gene function and lead to the development of various genetic disorders. For example— Huntington’s disease.
In the case HD, an expansion of CAG repeats within the HTT gene results in the production of a mutant huntingtin protein, leading to neurodegeneration and the characteristic symptoms of the disease.
Based on the number of repeats, however, the condition can be categorized into normal, premutation and full mutation (13). To know more, you can read our entire series of articles on this topic.
Topic: Triple Repeat Expansion.
Mitochondrial Mutations:
Our cells contain extracellular or cytoplasmic DNA in the membrane-bound organelles like mitochondria. The mtDNA has its own replication and gene expression machinery. The usual function of mitochondria is to provide energy.
However, mutations in mitochondrial DNA can impact mitochondrial function and contribute to a variety of disorders affecting energy-dependent tissues. For example— mitochondrial myopathy.
The present condition is associated with impaired production of energy within muscle cells, leading to muscle weakness, fatigue, and other symptoms (14).
Related article: Organelle DNA- Mitochondrial and Chloroplast DNA.
Transposon Activities:
Popularly known as “jumping genes,” transposons or transposable elements have the power to change their position within the genome. They literally can jump between gene locations in the genome, which when occurring within a gene, causes faults in the gene product.
Put simply, When transposons move to new locations, they can disrupt normal gene function or regulatory elements, potentially leading to genetic disorders. Such transposition indirectly produces genomic instability and contributes to conditions such as cancer and neurological disorders.
For example— the transposition of Alu elements (15). Notedly, transposons played a pivotal role in evolution too. Read this article to learn more.
Read more: Role of Transposons in Evolution of Eukaryotic Genome.
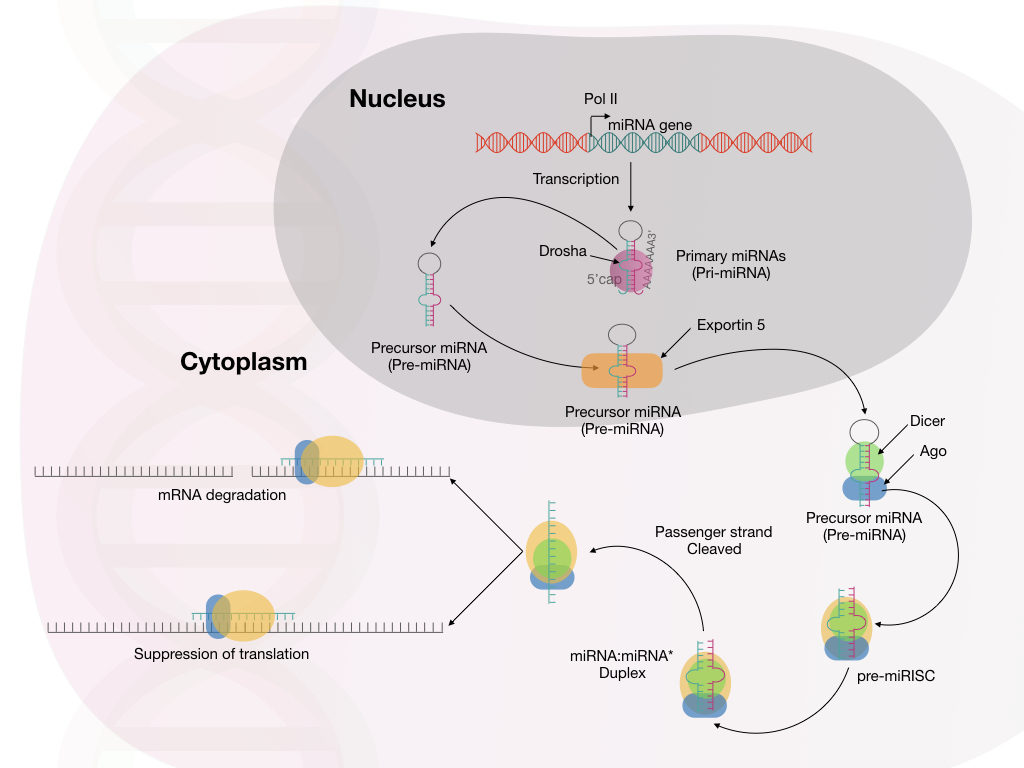
Non-coding RNA Dysregulation:
The specific role allotted to the coding mRNA is to synthesize the protein. However, not all the RNA can participate in the present process. Such undestined RNAs are non-coding RNAs.
microRNA and long non-coding RNAs are common non-coding RNAs, that play critical roles in gene expression regulation. Dysregulation of non-coding RNA-mediated gene expression leads to abnormal cellular function and disease conditions. For example microRNA-21 mediated cancer.
Dysregulation of microRNA-21 has been known in several cancers. Increased expression of miR-21 is associated with cellular dysregulation and spread of cancer (16).
Copy Neutral Loss of Heterozygosity (CN-LOH):
CN-LOH is yet another interesting genetic phenomenon. Here the loss of the one copy doesn’t affect the number of copies of that particular gene.
Meaning, that, Copy-Neutral Loss of Heterozygosity (CN-LOH) involves the loss of one of the two alleles without any change in copy number. The present condition is difficult to determine due to the balanced copy numbers.
However, the CN-LOH can result in the unmasking of recessive mutations and contribute to genetic disorders. The present condition is reported in certain cancers.
For example in CN-LOH-mediated leukemia, the loss of heterozygosity in specific genomic regions is reported (17).
Related article: What is Loss of Heterozygosity? How It is Associated with Cancer?
Errors during DNA Metabolisms:
Replication, transcription, translation, post-translational modification and DNA repair are DNA metabolism events that occur in sync. However, mistakes in any of these processes can produce serious health-related complications.
For example, errors in the DNA repair pathway produce genomic instability and result in cancer-like conditions.
BRCA-mediated hereditary breast cancer syndrome is the best example that explains the present scenario. Here, defective DNA repair mechanisms cause hereditary breast cancer syndrome and increase the risk of breast and ovarian cancer (18).
Wrapping up:
Genetic disorders can be caused by a variety of factors or a combination of many different factors, but these 15 mechanisms are the most common. Keep in mind that inherited genetic conditions are passed on to the next generation without any known reasons.
Because either parent has the condition, the offspring may likely get it!
So that’s it for this article. I strongly believe that this article must add value to your genetic knowledge. Do share this article. And if you wish you can enroll in our genetic academy.
Sources:
- Genetic disorders and congenital abnormalities: strategies for reducing the burden in the Region by WHO.
- Rare Genetic Disease By National Human Genome Research Institute.
- Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015 Jan;16(1):45-56. doi: 10.1038/nrg3849.
- Prakash J, Mittal B, Srivastava A, Awasthi S, Srivastava N. Association of FTO rs9939609 SNP with Obesity and Obesity- Associated Phenotypes in a North Indian Population. Oman Med J. 2016 Mar;31(2):99-106. doi: 10.5001/omj.2016.20.
- Catović A, Kendić S. Cytogenetic findings at Down syndrome and their correlation with clinical findings. Bosn J Basic Med Sci. 2005 Nov;5(4):61-7. doi: 10.17305/bjbms.2005.3236.
- Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, Liu B, Feng JX, Pan YJ, Yan JS, Liu Q. The Philadelphia chromosome in leukemogenesis. Chin J Cancer. 2016 May 27;35:48. doi: 10.1186/s40880-016-0108-0.
- Taylor CM, Smith R, Lehman C, et al. 16p11.2 Recurrent Deletion. 2009 Sep 22 [Updated 2021 Oct 28]. In: Adam MP, Feldman J, Mirzaa GM, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2023.
- Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010 Dec;23(4):213-27. doi: 10.1177/0891988710383571.
- Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015 Jan 22;16(2):2472-96. doi: 10.3390/ijms16022472.
- Satizabal CL, Samieri C, Davis-Plourde KL, Voetsch B, Aparicio HJ, Pase MP, Romero JR, Helmer C, Vasan RS, Kase CS, Debette S, Beiser AS, Seshadri S. APOE and the Association of Fatty Acids With the Risk of Stroke, Coronary Heart Disease, and Mortality. Stroke. 2018 Dec;49(12):2822-2829. doi: 10.1161/STROKEAHA.118.022132.
- Duraturo F, Liccardo R, De Rosa M, Izzo P. Genetics, diagnosis and treatment of Lynch syndrome: Old lessons and current challenges. Oncol Lett. 2019 Mar;17(3):3048-3054. doi: 10.3892/ol.2019.9945.
- Ma VK, Mao R, Toth JN, Fulmer ML, Egense AS, Shankar SP. Prader-Willi and Angelman Syndromes: Mechanisms and Management. Appl Clin Genet. 2023 Apr 6;16:41-52. doi: 10.2147/TACG.S372708.
- Nopoulos PC. Huntington disease: a single-gene degenerative disorder of the striatum. Dialogues Clin Neurosci. 2016 Mar;18(1):91-8. doi: 10.31887/DCNS.2016.18.1/pnopoulos.
- Ahuja AS. Understanding mitochondrial myopathies: a review. PeerJ. 2018 May 21;6:e4790. doi: 10.7717/peerj.4790.
- Zhang W, Edwards A, Fan W, Deininger P, Zhang K. Alu distribution and mutation types of cancer genes. BMC Genomics. 2011 Mar 23;12:157. doi: 10.1186/1471-2164-12-157.
- Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016 Oct;5(4):395-402. doi: 10.3892/br.2016.747. Epub 2016 Aug 26.
- Mullighan CG, Downing JR. Global genomic characterization of acute lymphoblastic leukemia. Semin Hematol. 2009 Jan;46(1):3-15. doi: 10.1053/j.seminhematol.2008.09.005.
- Shiovitz S, Korde LA. Genetics of breast cancer: a topic in evolution. Ann Oncol. 2015 Jul;26(7):1291-9. doi: 10.1093/annonc/mdv022. Epub 2015 Jan 20.

you are in reality a just right webmaster The site loading velocity is incredible It seems that you are doing any unique trick In addition The contents are masterwork you have performed a wonderful task on this topic