“By partitioning the sample into smaller volumes, absolute quantification of nucleic acid is performed with high accuracy and precision using digital PCR.”
The foremost application of PCR is to either amplify the DNA or quantify it or both in the same reaction.
In the conventional PCR, the DNA of our interest amplified for getting multiple copies of it because single or couple of DNA molecules are not sufficient for downstream assays.
However, gene quantification is not possible using conventional PCR, thus, gene expression and nucleic acid quantification can not be done by conventional PCR.
For quantification of nucleic acid, viral load and pathogen, we need an advanced PCR machine called real-time thermocycler.
Using the fluorescent chemistry the amount of nucleic acid present in a sample can be measured by quantitative PCR.
Using either fluorescent dye or hydrolysis probe, the relative quantification can be done in realtime PCR for gene expression, microRNA studies and pathogen load quantification.
Read our mammoth article on realtime PCR: Real-time PCR: Principle, Procedure, Advantages, Limitations and Applications.
The realtime PCR is now a gold standard method for quantification of cytokines and even, it is the most trusted quantification machine for TB detection.
Albeit, due to several limitations of realtime PCR we need an advanced, robust and more accurate method for quantification of nucleic acid.
In the present article, we are going to discuss digital PCR, an advanced method for quantification of nucleic acid. The content of the article is,
Read further: The Polymerase Chain Reaction.
Key Topics:
Overall idea:
Using realtime PCR scientists can estimate the amount of DNA or RNA present in a sample. However, the large volume used during reaction preparation can lead quantification of unwanted nucleic acids too.
In addition to this, several molecules are not quantified, although the redundancy is not so large.
To overcome this problem, digital PCR is invented.
The principle of digital PCR is based on the absolute quantification of a sample by dividing it into smaller micro-volumes.
The overall idea of dividing the sample into smaller volumes is to quantify a single molecule present in that micro-volume, thus increases the precision of quantification.
What is digital PCR?
The digital PCR often known as dPCR or dePCR is used for absolute quantification of gDNA, cDNA or RNA.
In the year, 1999, Bert Vogelstein and Kenneth Kinzler discovered “digital PCR” by analysing the rare genotypes of cancer mutations. Although the concept of digital PCR is developed in 1992 by Sykes et al.
The method is employed frequently for detection of rare alleles and detecting point mutations.
Furthermore, it is used in clonal amplification during NGS.
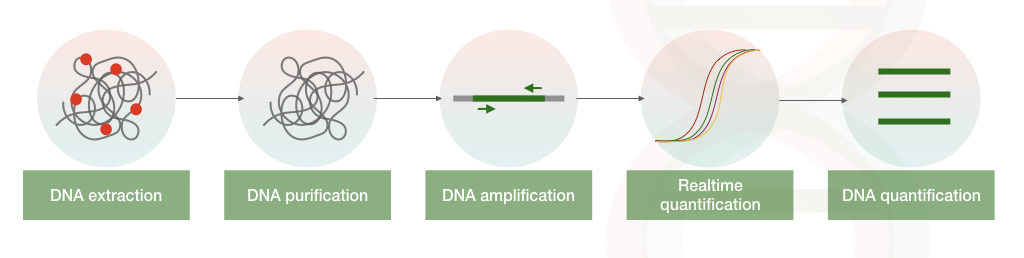
The current gold standard method for quantification of nucleic acid is realtime PCR which relies on the fluorescent chemistry.
Either hydrolysis-based fluorescent probe or DNA intercalating dye is used in the quantification of nucleic acid.
In a nutshell, once the fluorescently labelled probe finds its complementary strand in a sample it binds to the sequence, the hydrolysed probe emits fluorescent which is detected by the machine.
In the intercalating dye-based method, the fluorescent dye binds to only the dsDNA and emits fluorescent. The amount of fluorescent emitted is measured by the machine.
However, “conventional qPCR” application required standards.
Standards are used as a baseline for measuring the sample, also, it is required in the same amount of the sample volume.
In addition to this, performing multiplexing quantification assay required expertise. Performing multiplexing assays is not an easy task due to the competitive assay nature.
The realtime PCR is often not applicable in the detection of rare alleles or heterogeneous samples because the limit of quantification between two samples is 2 fold.
These problems are overcome by using the new generation nucleic acid quantification method called digital PCR which counts the total number of DNA molecules in digital formate by dividing a sample in micro-volumes.
The workflow of the digital PCR is alike the qPCR utilises same reagents and similar thermocycler conditions.
The chemistry is based on the use of fluorescent molecules for detection (same as qPCR). dNTPs, primer set, PCR reaction buffer, template DNA, Taq DNA polymerase and other PCR enhancers are used in PCR reaction preparation.
Note:
We have to use the hot-start DNA polymerase in every quantification PCR assays. Hotstart Taq DNA polymerase decreases the chance of non-specific amplification.
Principle of digital PCR:
In the quantitative PCR, the initial PCR cycles are not exponential thus, the nucleic acid can not be quantified accurately.
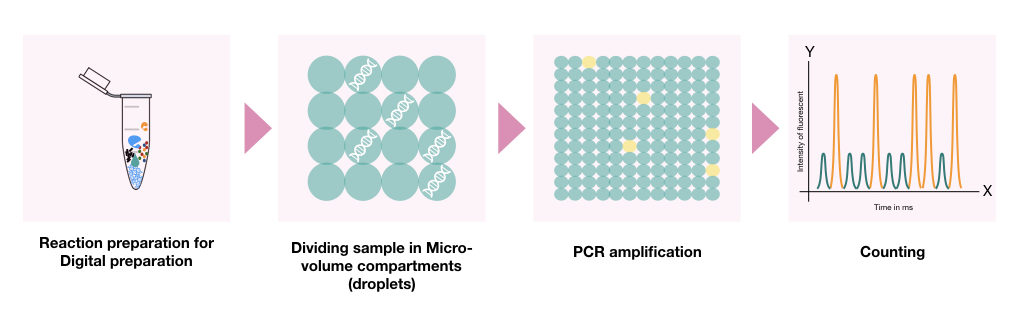
In the dPCR, once the PCR reaction mixture is prepared, the PCR reaction mixture (along with the template DNA) partitioned into thousands of smaller droplets in which separate PCR reaction occurs in individual droplets.
(PCR reaction for the droplet PCR: dNTPs, hot-start DNA polymerase, MgCl2, optimum reaction buffer, primers, fluorescent probe and quencher probes).
The mode of processing is similar to the realtime PCR, once the fluorescently labelled probe binds to the DNA it emits the light measured by the machine.
In each micro-volume reaction (droplet), amount of amplification and numbers of nucleic acid molecules are measured using the Poisson distribution method.
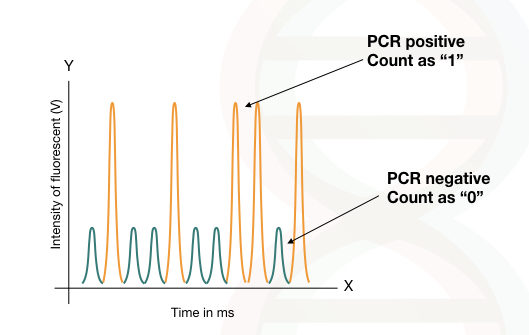
The fluorescence emitted by the droplet with no template is called as “ 0” and it is used as a baseline or as the zero for the calculation, (PCR-negative reaction).
The amount of the fluorescent emitted by the droplet with a single template is called “1” and recorded as one, (PCR-positive reaction).
The absolute quantification of target DNA is done using the Poisson distribution method using the numbers of positive vs negative reaction done in each droplet.
Once the PCR reaction partitioning is done, the polymerisation is progressed to its endpoint.
The reaction droplet generator machine divides a sample in more than 20,000 nano-litter sized droplets.
Thus, no reference, calibration curve or endogenous control required to quantify DNA.
In the dPCR, the reaction mixture is divided into smaller micro-volume reactions in such a way that only one or zero target molecule may present per micro-volume reaction.
In the final calculation, the total number of positive reactions (having 1 template) is equal to the total number of target DNA present in the original volume of a sample.
The absolute concentration of total volume is calculated using this equation:
Absolute concentration = a total number of template DNA counted / total volume measured in each droplet (or well or micro-volume reaction).
Note:
The dPCR method is based on the TaqMan chemistry in which the fluorescent-dye labelled probe is used complementary to the template.
Dye FAM (fluorescent dye) is used along with the HEX reporter dye.
Methods for creating sample partitions: Oil emulsion, microwell plates, nono fluid chip and capillaries.
Applications of digital PCR:
- Rare allele detection As well as rare sequence detection.
- Accurate copy number variation studies
- Accurate NGS library quantification
- Viral load detection
- Pathogen detection
- Single-cell gene expression assay
- Validation of low-frequency mutations identified by sequencing.
- Gene expression and microRNA research
Read our article on microRNA: microRNA (miRNA) and Gene Regulation.
One of the biggest advantages of dPCR:
The digital PCR can detect the difference in the gene expression of less than 30%, only a single copy number variation can be distinguished accurately.
In addition to this, it can identify the alleles having allelic frequencies less than 0.1%.
Advantages:
- Simple set up not required higher expertise as like in quantitative PCR.
- Higher sensitivity
- Higher reproducibility
- Quantify target in low samples
- Improved multiplexing capacity
- High tolerance capacity against PCR inhibitors
- Inear and accurate detection of low-frequency genotypes.
Droplet digital PCR:
Through the process of water-oil emulsion technique, the nano-volume sized oil droplet is created for partitioning the reaction.
The quantification of DNA using this technique is called droplet digital PCR. Although, the basic principle of the droplet digital PCR is similar to the dPCR.
The ddPCR generates more than 20,000 micro-volume partitions for absolute quantification.
“Higher the partitions higher the accuracy.”
If the reaction is partitioned is larger numbers of micro-volume reactions, the chance of having zero or one template in micro-volume reaction compartment is higher, thus, counting becomes more accurate.
Thus, ddPCR (using water-oil emulsion technique) is advisable for rare allele and low-frequency genotype detection.
Read more: DNA Sequencing: History, Steps, Methods, Applications And Limitations.
Conclusion:
Digital PCR is one of the most accurate methods used for quantification of nucleic acid. Its power of detecting even a single allele makes it a bigger player in cancer research and detection of rare mutations and rare alleles in developing cancer.
Sources:
- Kanagal-Shamanna R. (2016) Digital PCR: Principles and Applications. In: Luthra R., Singh R., Patel K. (eds) Clinical Applications of PCR. Methods in Molecular Biology, vol 1392. Humana Press, New York, NY. doi: 10.1007/978-1-4939-3360-0_5.
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96(16):9236–9241. doi:10.1073/pnas.96.16.9236.


