“Short-read sequencing can sequence 50 to 300 bp shorter DNA fragments effectively. Explore the concept of short read sequencing and learn about its importance, advantages, applications and limitations.”
In the previous article, we explained sequencing reads in NGS. Put simply, it’s the length of the nucleotide fragment going to be sequenced in the machine. Short and long-read sequencing are two of its types.
Short-read sequencing can sequence shorter DNA fragments of 50 to 300 bp while long-read sequencing can sequence longer DNA fragments of 1000 to 100,000 bp. Both have their own importance and limitations.
However, short-read sequencing is extensively employed in clinical studies and diagnosis, and thus the vastly adopted chemistry in most NGS platforms is short read sequencing.
In this article, I will explain to you the concept of short read sequencing, how it’s done and why it is important.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is Short-Read Sequencing?
Short-read sequencing is a next-generation sequencing chemistry in which shorter DNA fragments are sequenced effectively. Usually, fragment lengths between 50 to 300 bp or shorter than 1kb (in general) can be considered as short reads.
Notedly, an increment in read length decreases the efficiency and sequencing accuracy. Shorter fragments are easy to generate, amplify and sequence, and hence excellent accuracy, efficiency and precision can be achieved.
The concept of short-read sequencing was inspired by the performance of Sanger sequencing. Researchers noted that Sanger sequencing could effectively sequence sequences a few hundred base pairs long.
They realized that to achieve rapid, precise, and accurate whole-genome sequencing, they needed to adopt the short-read sequencing approach, which eliminates the need for post-sequencing trimming.
Why is short read sequencing important?
Our genome is extremely complex and huge. It’s nearly impossible to sequence the whole genome in a single run or in a few larger chunks. Third-generation long-read sequencing such as PacBio HiFi or Oxford Nanopore technology, although can sequence longer DNA fragments, has its own limitations.
Long read sequencing is an inaccurate, time-consuming and costly process. As a result, short-read sequencing has become the standard chemistry for NGS platforms.
Short read sequencing is fast, accurate, cost-effective and can sequence the whole genome in a day (roughly). Therefore, it’s important for clinical and diagnostic setups.
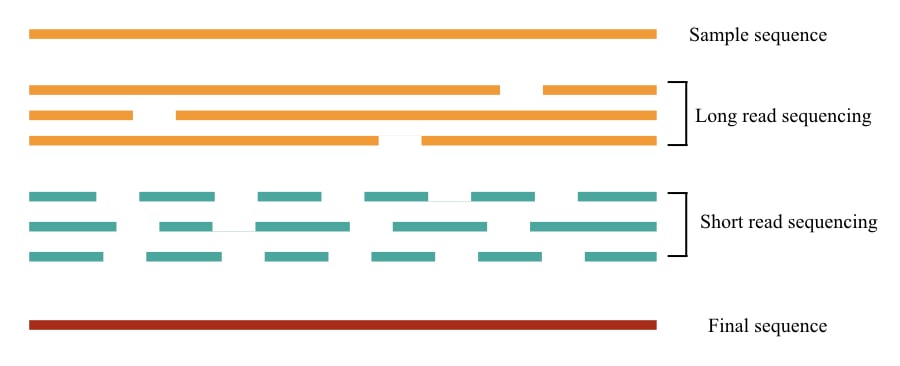
Types of Short-Read Sequencing:
As aforementioned, short read sequencing has been widely accepted chemistry in various NGS platforms. Illumina is the prime player in this segment with its HighSeq series, MiSeq and NextSeq series.
Followed by Illumina, Thermo Fisher Scientific’s Ion Torrent and Ion proton also use short read sequencing. In addition, PacBio’s ONSO system also uses short read sequencing chemistry.
However, each platform has its own chemistry for whole genome sequencing. Let’s understand the chemistry behind each type of short read sequencing platform.
Short read sequencing chemistry
In this section, we will elaboratively understand the chemistry of each short read sequencing platform.
Sequencing by synthesis
Sequencing by synthesis is a unique chemistry in which a sequence has been read during the synthesis process. Meaning, during the incorporation of nucleotides. A fluorescence signal, generated during the addition of each nucleotide is read by the machine and converted into digital signals.
Here, a short DNA fragment of 150 bp is ligated with the adaptors, index sequences and flow cell-specific primers. Now, the fragment library is added to the flow cell and allowed for sequencing by synthesis.
Illumina’s instruments HiSeq, MiSeq and NextSeq series work on the principle of single or paired-end short read sequencing by synthesis chemistry. Learn more about it here: What is Sequencing by Synthesis?
Semiconductor sequencing
In this sequencing chemistry, short DNA fragments are read by the detection of hydrogen ions. During the nucleotide incorporation, when the polymerase adds nucleotides, hydrogen ions are released.
This event leads to a change in the pH, the semiconductor measures the pH change and converts it into sequence information. Millions of such changes are measured simultaneously for a particular DNA fragment or sequence.
Thermo Fisher’s Ion Torrent and Ion Proton and other NGS machines work on the principle of semiconductor sequencing.
PacBio’s sequencing by binding also works on short read sequencing (fragment size between 150 to 217). In this chemistry, the short fragments are read by incorporation of labeled nucleotides on to the target fragment.
Short read sequencing process:
The short read sequencing process is similar to other NGS platforms. Here is a short summary of the process.
| Step | Explanation |
| Nucleic acid isolation | High-quality DNA or RNA is isolated using a kit or automated DNA extractor. |
| Fragmentation and library preparation | The genomic DNA is fragmented and the fragment library is prepared during the library preparation procedure or in the instrument. |
| Sequencing | Based on the chemistry of the machine, fragments are sequenced in the machine. |
| Quality control | Read quality assessment is performed to collect the qualified reads. |
| Read alignments | Now, the short reads are aligned with the reference genome. |
| Variant calling | Variations from the target sequence are noted. |
| Variant annotation | Variations are annotated using software. |
Advantages and Applications:
Short-read sequencing offers high accuracy, rapidity, and cost-effectiveness. It is supported by well-established bioinformatics tools and pipelines, making it widely applicable in various assays, including whole-genome sequencing (WGS), whole-exome sequencing (WES), RNA sequencing, and chromatin immunoprecipitation sequencing (ChIP-seq).
It’s also important to quote that the short read chemistry can sequence the low-quality DNA samples, precisely.
Whole-Genome sequencing
The present chemistry has been extensively utilized in genome-wide studies. The entire genome is fragmented into smaller-sized fragments, sequenced, read and analyzed using bioinformatic tools.
Genome-wide sequencing helps study genome-wide alterations and identify novel variations present in the genome, particularly associated with a disease.
Whole-Exome sequencing
Short read sequencing is also used to study only the coding sequences present in the genome to find out any alterations present in the coding regions of genes. The present technique helps to identify alterations associated with protein-coding sequences.
Metagenomic studies
Short-read NGS chemistry is extensively utilized for metagenomic studies as well. It helps effectively study the DNA sequences from thousands to millions of microbes present in the sample.
Related article: What is Metagenomics?- Definition, Steps, Process and Applications.
RNA-sequencing
Short read sequencing is also a preferred technique for transcriptomics and gene expression studies.
Clinical diagnosis
Due to the higher accuracy, lower cost and TAT, short read sequencing is extensively applied in disease diagnosis. For example, genome, exome or transcriptomic sequencing helps find genomic variations associated with a particular disease.
In the diagnostic industry, short-read sequencing finds extensive application in cancer diagnostics, reproductive biology, and pediatric fields.
Other than that, short read sequencing is also used in targeted sequencing, gene panel sequencing and pharmacogenomics and nutrigenomics.
Limitations:
Despite having extensive applications in various fields, short read sequencing has several limitations as well.
It can not investigate complex genomic regions such as repetitive and high GC-rich regions.
It can not investigate complex and large genomic alterations such as inversions, deletions or duplications.
It also can not perform phasing haplotyping effectively.
Uneven coverage presents another limitation of current sequencing chemistry. Certain genomic regions may be over-sequenced, while others are under-sequenced, resulting in ambiguity in the obtained results.
Studying short read sequencing data is bioinformatically challenging. Data generated during the short read sequencing are 50 GB to 1TB which is difficult to process and study.
Related articles:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- What is NGS?- Definition, Principle, Steps, Chemistries, Advantages and Limitations
- De Novo Sequencing: Steps, Procedure, Advantages, Limitations and Applications
Wrapping up:
Short read sequencing, as I said above, is the core chemistry of NGS. It’s cost-effective, fast and reliable, making it widely used in research and medical diagnoses. However, it does have its drawbacks.
It’s a first choice for genome, exome and transcriptome sequencing. So, utilized for GWAS, WES, target sequencing, metagenomics, pharmacogenomics, nutrigenomics and cancer research.
I hope this article will help you to learn sequencing more effectively. Do share the article and bookmark the page.
Sources:
Simon SA, Zhai J, Nandety RS, McCormick KP, Zeng J, Mejia D, Meyers BC. Short-read sequencing technologies for transcriptional analyses. Annu Rev Plant Biol. 2009;60:305-33. doi: 10.1146/annurev.arplant.
Short read sequencing by Genomic notes for clinicians.
Short read sequencing vs long read sequencing by Sampled.


