“The Cas9 protein is a nuclease that destroys phage DNA using the double RNA-guided DNA cleavage through DNA binding and nuclease activities.”
Key points:
- Cas9 protein is predominantly present in type II bacterial CRISPR systems.
- It needs both crRNA and tracrRNA for effective working.
- It also requires a PAM sequence on the target DNA for catalytic activity.
- Various modified Cas9 are used for different purposes ranging from activating genes to suppressing gene expressions.
Cas9 has been known for its significant importance in CRISPR mediated gene editing and applications like disease modeling, studying the role of genes, therapeutic and gene expression studies.
Put simply, it finds, binds and cleaves the target nucleic acid using the PAM sequence as a marker. It employs the sgRNA having cRNA and tracrRNA and finds complementation with the target location to determine the fugitive.
However, its two-level authentication (the use of sgRNA & PAM) greatly reduces efficacy during in vitro gene editing. So customized Cas9 nucleases like the spCas9, dCas9, SaCas9, XCas9 etc are available.
In the present article, I will explain various types of Cas9 nuclease, structure and function of each. I will also try to explain the importance of various Cas9 proteins for gene therapy.
Stay tuned,
Explore the whole category: CRISPR-Cas9.
Key Topics:
What is Cas9?
RNA-guided DNA endonuclease
Cas9 or CRISPR-associated protein 9 is one of the well-studied, important and commercially available nucleases used not only in the bacterial systems but also in in vitro gene-editing techniques.
Only present in CRISPR type II, the Cas9 is a type of DNA nuclease that can cleave off dsDNA precisely. It is often referred to as dual RNA guided DNA endonuclease, most commonly found in Streptococcus pyogenes.
To understand why only Cas9 is popularly used in gene editing, we have to understand the structure, function and importance of the Cas9 protein which was previously known as Cas5, Csx12 or Csn1.
| Name | Cas9 endonuclease |
| Alternative name | spCas9/spyCas9 |
| Organism | Streptococcus pyogenes serotype M1 |
| Molecular weight | ~163KDa |
| Gene | cas9 |
| Location on chromosome | 0.85 to 0.86Mb |
| Protein | CRISPR-associated endonuclease Cas9/Csn1 |
| Cofactor | Mg2+ |
| Biological processing | Interference- defense response to phage. Maintaining CRISPR repeat sequences |
| Functions | DNA and RNA binding Metal ion binding 3’-5’ exonuclease activity Endonuclease activity |
Structure of Cas9:
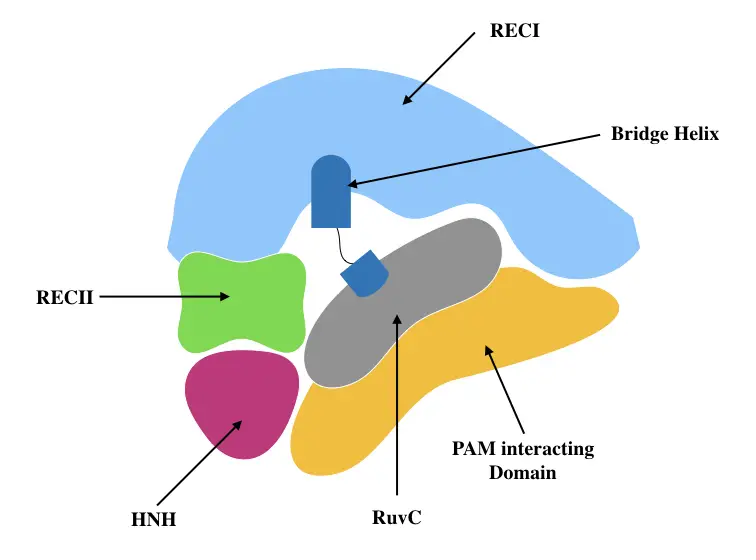
Crystallographic analysis reveals that the Cas9 protein is a bilobed structure, each lobe has various domains to execute a separate function. The two lobes are the Recognition lobe (REC) and the nuclease lobe (NUC).
As the names themselves suggest largely, the REC lobe recognizes the target DNA while the NUC lobe accomplishes catalytic activity thereby Cleavage of dsDNA.
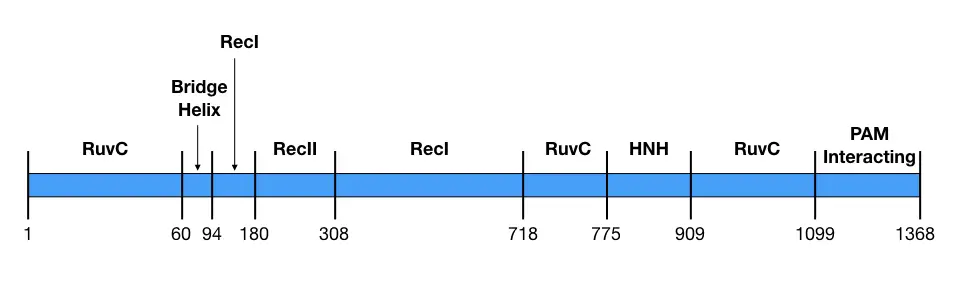
Comprehensive analysis showed that the REC lobe has three separate domains viz alpha-helix or Bridge helix, REC1 domain and REC2 domain while the NUC lobe has RuvC, HNH and PAM- interacting domain (Nishimasu et al., 2014). The structure is pictorially shown here.
Note
The Arginine-rich region in the structure of Cas9 is highly conserved and has DNA binding activities, research suggests.
A detailed explanation of each domain is discussed here:
| Lobe | Domain | Residues | function |
| REC | Bridge helix | 60-93 | Recognition of DNA |
| REC | REC1 | 94-179, 308-713 | RNA guided DNA targeting |
| REC | REC2 | 180-307 | DNA binding |
| NUC | RuvC (RuvCI, RuvCII and RuvCIII) | 1-59, 718-769, 909-1098 | RNase H activity; Nuclease activity for non-complementary target strand. |
| NUC | HNH | 775-908 | Nuclease activity for complementary target strand |
| NUC | PAM-interacting- domain | 1099-1368 | Finds the PAM sequence on the target DNA. |
Oakes et al., 2014 explained that the structure of Cas9 is hand-shaped with 100 * 100 * 50Å size. Note that they have also reported a RuvC domain in the REC lobe. Put simply, the REC lobe finds the complementary DNA region while the NUC lobe cleaves it by dual RNA-guided dsDNA endonuclease activity. The structure was precisely explained by Doudna & Jiang, 2017.
Functions of the Cas9:
Although every domain has a unique function; largely, the overall function of Cas9 nuclease is as follows.
Preliminarily, the Cas9 recognizes the PAM sequence on the anti-target sequence, afterward, forms the Cas9-sgRNA binary complex to recognize the target DNA. It forms a tertiary (Cas9-sgRNA)-dsDNA complex by interaction with 20 complementary nucleotides of the DNA.
In the next step, the tertiary complex’s activity recognizes the nonself DNA and starts the catalytic reaction. Noteworthy, there are many different mechanisms bacteria use to identify self vs nonself DNA and is vary among different systems.
In the final stage, it cleaves the dsDNA and completes the interference process.
Note that, though the Cas9 is involved in DNA and RNA target processing, it only stabilizes the crRNA-tracrRNA complex during the RNA process but doesn’t execute the catalytic activity. This owing that the catalytic nuclease activity is only restricted to DNA processing only!
However, the PAM sequence is necessary to activate Cas9 and distinguish self vs non-self DNA. The Cas9 can’t do its function if it does not find the PAM sequence. If the PAM sequence mutates, the crRNA-tracrRNA complex can’t form.
In addition, it also needs Mg2+ ions as a cofactor for doing so. Various in vitro studies suggest that the nuclease or catalytic activity can be suppressed or controlled by adding the EDTA. Here I have listed several species and their known PAM sequence.
| Cas9 nuclease | Species | PAM sequence requirement (3’ to 5’) |
| SpCas9 | Streptococcus pyogenes | NGG |
| SaCas9 | Streptococcus aureus | NGRRT or NGRRN |
| CjCas9 | Campylobacter jejuni | NNNNRYAC |
| StCas9 | Streptococcus thermophilus | NNAGAAW |
| TdCas9 | Treponema denticola | NAAAAC |
(The information of the table derived from the addgene blog)
Mechanism of working:
During the process of bacterial interference, in the very first step, the ribonucleoprotein complex (RNP) is formed by the interaction between the REC lob and the gRNA complex, consecutively, nuclease domain RuvC and HNH hydrolyze two phosphodiester bonds of two separate strands resulting in dsDNA strand separation.
An in-depth study signifies that the HNH active domain hydrolyzes the phosphodiester bond of the complementary strand while the RuvC active site hydrolyzes the phosphodiester bond of the non-complementary strand. As it requires metal ions for activation, RuvC and HNH use two-metal ions and one-metal ions, respectively for hydrolysis (Tang H et al., 2021).
Different types of Cas9 nucleases:
Various types of natural and artificially occurring Cas9 nucleases exist which are categorized depending upon either their function or species they derived. Here I am enlisting and explaining some of them.
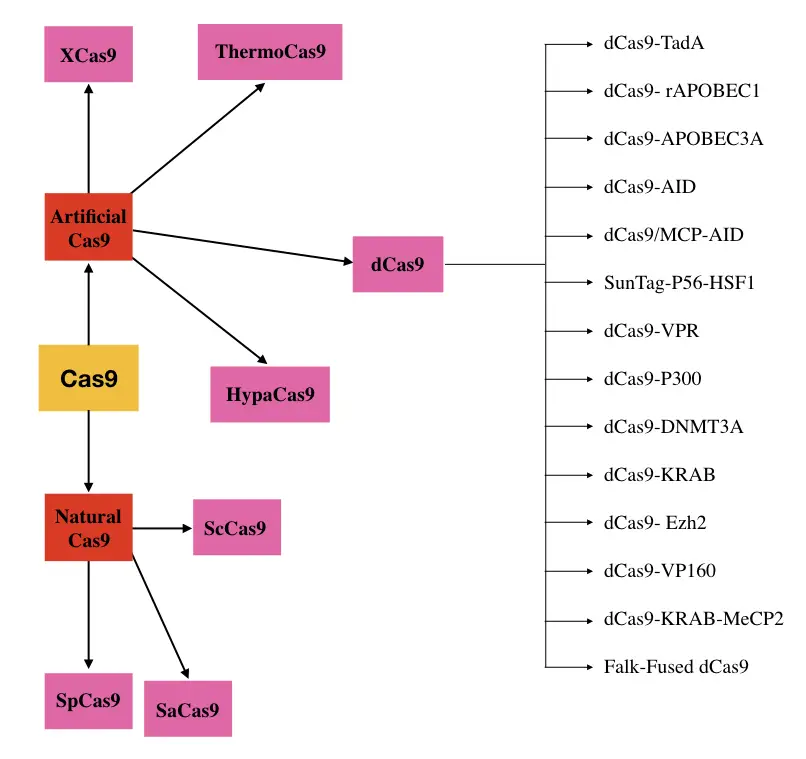
SpCa9:
The SpCas9 is derived from the Streptococcus Pyogenes and is one of the most popular, well-studied and widely-used Cas9 nucleases in genetic engineering experiments. As aforestated, It requires both crRNA and tracrRNA as sgRNA as well as PAM sequence to identify the target.
Once the SpCas9 finds the PAM (5’-NGG-3’) sequence, the sgRNA immediately locates the nuclease on the target region where the spCas9 performs a double-stranded cut.
The structure is similar to the abovementioned general structure of the Cas9 with the nuclease lobe for catalytic activity and recognition lobe for recognizing and identifying the target DNA.
(Here I am not repeating things but giving a broad overview in a tabular representation.)
| Structure | Bilobed (REC and NUC) |
| Domains | NUC (Nuclease domain): HNH and RuvC REC (recognition domain): Rec1, Rec2 and Rec3. |
| Bacterial CRISPR system | System II |
| PAM sequence | 5’-NGG-3’ (N is any nucleotide) |
| SgRNA | Required (crRNA: tracrRNA) |
| Variants | SpCas9-NRRH, SpG, SpCas9-NRCH, SpCas9-NRTH, |
| Key points (Advantages) | Shortcomings (disadvantages) |
| Easily available and well-studied. Easy to isolate Highly effective Easily to use | Required PAM sequence. Also finds false PAM and produces off-target effects. Recognize other PAMs like 5’-NAG-3’ or 5’-NGA-3’. Large in size and can’t be delivered easily. Difficult to deliver and express. |
Applications:
As aforementioned, the present system has been precisely studied, having huge data and hence popular in gene therapy. Several common applications are
- Transcriptional repression
- Transcriptional activation
- Epigenetic modulation
- Gene disruption
- Single-base pair conversion
To learn more on different applications of CRISPR cas9, read this article: Applications of CRISPR-CAS9 in Medical Science, Diagnostics, Research, Plant Biology and Agriculture.
SaCas9:
Yet another most popular Cas9 nuclease is the SaCas9 which is though structurally similar to the SpCas9, to a lesser extent; but differs in terms of size. The compact size of SaCas9 is its admirable property. Henceforth, it is a suitable replacement for the SpCas9.
SaCas9 is derived from the Streptococcus aureus having only 1053 amino acids in its structure which is about 1Kb smaller than the SpCas9.
It also needs a PAM sequence as 3’-NNGRRT-5’ to distinguish self vs non-self DNA. Upon catalysis, it generates sticky double-stranded ends.
| Structure | Bilobed (REC and NUC) |
| Domains | NUC (Nuclease domain): HNH and RuvCREC (recognition domain): Rec1, Rec2 and Rec3. |
| Bacterial CRISPR system | System II |
| PAM sequence | 5’-NNGRRT-3’ (N is any nucleotide) |
| SgRNA | Required (crRNA: tracrRNA) |
| Variants | efSaCas9, KKHSaCas9 and SaCas9-HF |
| Key points (Advantages) | Shortcomings (disadvantages) |
| Small in size Great precision Versatile Accurate Easily packed into a viral vector | Required PAM sequence Need a larger sgRNA to work substantial off-target effect |
Applications:
The present Cas9 nuclease is widely used in plant genome editing in
- Plant-pathogen interaction studies
- Stress tolerance studies
- Pathogen resistance investigations
- It can also be used in treating infectious and genetic diseases.
Note
A specialized SpCas9 is recently used to determine the role of the Myostatin gene in Muscular atrophy.
ScCas9:
The ScCas9 nuclease is isolated from Streptococcus canis which required slightly a different PAM recognition site for its action which is 5’-NNG-3’ (instead of NGG). The present nuclease is structurally similar to other Cas9, however, due to its less effectiveness, it isn’t advisable.
| Name | ScCas9 |
| Species derived | Streptococcus canis |
| PAM sequence | 5’-NNG-3’ |
| sgRNA requirement | Yes, as crRNA:tracrRNA |
| Variants | SpCas9++, SpCas9n++ |
ScCas9 and its variants like SpCas9++, SpCas9n++ and SpCas9+ are popularly used in plant genome editing.
dCas9:
dCas9 is one of the most sophisticated, versatile, amazing and out-of-the-box variants of the Cas9 nuclease, why? Because it lacks the primitive function of the nuclease which is “nucleolytic activity”. Therefore, it is known as the dead Cas9 system.
After removal of the catalytic domain, the remaining recognition domains only find the target DNA but do not cut it. So technically, various transcriptional factors can be transferred to a target location (Isn’t it amazing!).
Do you know?
Qi et al., in 2013 has created the first Dead Cas9 system.
We have written a whole article on this topic, if you wish to learn more on dead Cas9, you can read it here: dCAS9 (Dead CAS system): Concept, Functions and Applications.
As it has tremendous popularity, various variants exist for different experiments, several common variants of dCas9 are enlisted here:
| dCas9 variant | Function |
| dCas9-TadA | repair mutated resistance in gene bacteria, preserve adenosine deaminase activity. The present modification is capable enough to repair the faulty or mutated resistance gene for various gene editing purposes. |
| dCas9-rAPOBEC1 | preserves cytidine deaminase activity |
| dCas9-APOBEC3A | preserves cytidine deaminase activity |
| dCas9-AID | preserves cytidine deaminase activity |
| SunTag-VP64 | transcriptional activator used to study the effect of overexpression. |
| dCas9-VPR | tripartite complex and transcription activator |
| dCas9-CBP | rearranging chromatin structure by histone acetyltransferase domain. |
| Falk-fused dCas9 | transcriptional activator module |
ThermoCas9:
Mougiakos et al., (2017) engineered a specialized thermoCas9 nuclease that could work at a higher temperature, efficiently. It is isolated from Geobacillus thermodenitrificans T12 thermostable bacterium.
They also have explained that it is capable of gene deletion and transcriptional silencing even at higher temperatures (55℃) without compromising the sensitivity and PAM requirement. Largely, it works efficiently between temperatures 20 to 70℃.
It is also known as GeoCas9.
| SpCas9 | GeoCas9 | |
| Size | 1368AA | 1087AA |
| PAM | NGG | CRAA (R=A or G) |
| Spacer length | 20nt | 22nt |
| Temperature | 33-45 | 50-70 |
HypaCas9:
The HypaCas9 is a Hyper Cas9, which facilitates greater genome-wide specificity without decreasing the target activity in humans and mice cells. It also minimizes off-target activity. Technically, HypaCas9 is prepared by introducing N692A, M694A, Q695A and H698A mutation in the Cas9.
eSpCas9:
Enhanced specificity Cas9 is a mutant variant of the native SpCas9, reduced off-target activity through a single point mutation. It is also known as high fidelity spCas9 or highly specific Cas9.
XCas9:
The XCas9 is a specialized, genetically engineered nuclease that has decreased off-target effect with non-NGG PAM as well as NGG PAM. As we know, Cas9 needs a PAM sequence to work efficiently which increases the specificity and also produces noticeable difficulties in experiments.
XCas9 can recognize various PAM sequences like NGG, GAA and GAT and work effectively as well. Therefore it becomes more effective and efficient than SpCas9 or SaCas9 and minimizes the requirement of PAM to a greater extent (Hu et al., 2018).
Summary of the article: The table represents various types of Cas9 nuclease, origin, PAM sequence structure and specialization.
| Cas9 type | Origin | PAM sequence (5’ to 3’) | Specialization |
| SpCas9 | Streptococcus pyogenes | NGG | Cleaves dsDNA using the sgRNA |
| SaCas9 | Streptococcus aureus | NNGRRT or NNGRR(N) | Small off-targeting effect |
| ScCas9 | Streptococcus canis | NNG | The PAM sequence can be altered depending upon the variant used. |
| ThermoCas9 | Geobacillus thermodenitrificans T12 | CRAA (R=A or G) | Can work efficiently at a higher temperature. |
| StCas9 | Streptococcus thermophilus | NNAGAAW | High on-target cleavage activity |
| HypaCas9 | Streptococcus pyogenes | N/A | Greater genome-wide specificity |
| eSpCas9 | Streptococcus pyogenes | NGG | Enhanced SpCas9 work more effectively than native SpCas9 |
| NmCas9 | Neisseria meningitidis | NNNNGATT | Need longer cRNA which increases the accuracy |
| XCas9 | Streptococcus pyogenes | NGG and non-NGG | A specialized Cas9 that works with/without the PAM. |
| dCas9 | Streptococcus pyogenes | NGG | Specialized Cas9 that lacks nuclease activity |
| Cas9-DD | Streptococcus pyogenes | NGG | Destabilized Cas9 prepared to increase the accuracy and efficiency. |
| SpCas9-VQR | Streptococcus pyogenes | NGA | Altered PAM for increasing SpCas9 specificity |
| SpCas9-EQR | Streptococcus pyogenes | NGAG | Altered PAM for increasing SpCas9 specificity |
| SpCas9-VRER | Streptococcus pyogenes | NGCG | Altered PAM for increasing SpCas9 specificity |
| SpCas9-NG | Streptococcus pyogenes | NG | Altered PAM for increasing SpCas9 specificity |
| SpCas9-HF1 | Streptococcus pyogenes | NGG | Altered PAM for increasing SpCas9 specificity |
| evoCas9 | Streptococcus pyogenes | NGG | Altered PAM for increasing SpCas9 specificity |
| Sniper-Cas9 | Streptococcus pyogenes | NGG | Altered PAM for increasing SpCas9 specificity |
Data were derived from the addgene blog and Wada N et al., (2020).
Wrapping up:
Cas9 is very important for CRISPR-mediated gene editing, however, it should be modified depending upon the limitation or requirement of the experiment. CRISPR is such a technique that can only be experienced in the lab. But theoretical learning will surely increase the prior knowledge to get mastery.
It’s our small effort to provide available resources, knowledge and information regarding Cas9 protein. I can’t get all the information on all Cas9 proteins, I tried my level best to collect as much information as possible. I hope this article will surely help you.
FAQs:
Sources:
Nishimasu, Hiroshi et al. “Crystal structure of Cas9 in complex with guide RNA and target DNA.” Cell vol. 156,5 (2014): 935-49. doi:10.1016/j.cell.2014.02.001.
Oakes, Benjamin L et al. “Protein engineering of Cas9 for enhanced function.” Methods in enzymology vol. 546 (2014): 491-511. doi:10.1016/B978-0-12-801185-0.00024-6.
Zuo, Z., Liu, J. Structure and Dynamics of Cas9 HNH Domain Catalytic State. Sci Rep 7, 17271 (2017). https://doi.org/10.1038/s41598-017-17578-6
Mougiakos, I., Mohanraju, P., Bosma, E.F. et al. Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat Commun 8, 1647 (2017). https://doi.org/10.1038/s41467-017-01591-4
Hu, J., Miller, S., Geurts, M. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). https://doi.org/10.1038/nature26155.
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., & Lim, W. A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152(5), 1173–1183. https://doi.org/10.1016/j.cell.2013.02.022.
Wada, N., Ueta, R., Osakabe, Y. et al. Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol 20, 234 (2020).
Jiang, F., & Doudna, J. A. (2017). CRISPR-Cas9 Structures and Mechanisms. Annual review of biophysics, 46, 505–529. https://doi.org/10.1146/annurev-biophys-062215-010822.