“Multiplex PCR amplifies multiple DNA template regions, simultaneously using different sets of primers in a single PCR reaction.”
PCR technology has the best utility in research as well as in diagnostics. It amplifies and quantifies nucleic acid. A conventional/ old generation PCR can only amplify the DNA.
The Real-Time- PCR or the qPCR can not only amplify the DNA but also quantifies it. Meaning, we can get amplicons and the number of gene copies present in a sample.
PCR now has an advanced setup, more accurate, reliable and faster. Although previously, a separate reaction setup was required for different DNA samples.
Meaning, researchers have to perform separate reactions or place individual tubes for individual samples. That was a more time-consuming process. Moreover, the uniplex setup was a costlier and manpower-consuming process.
So anyhow scientists have to develop an assay or a modification in PCR that comprises multiple reactions in a single tube or reaction. Multiplex PCR has solved this problem- can save time and cost of the experiment.
Such techniques can change the game for diagnostics. To use a technique in the diagnostic industry, it should be low-cost, faster, less manpower-consuming, effective, efficient and most importantly accurate.
Multiplex PCR fulfills all the criteria listed above. What we are doing here is mixing several reactions in a single one, using almost the same quantity of reagents and a similar protocol.
Henceforth, the technique has serious applications in microbial genetic studies and gene quantification assays. Several SARS-CoV-2 COVID-19 detection techniques also rely on multiplex PCR.
Using multiple sets of primers in a single reaction, multiple template sites or different DNA regions can be amplified in multiplex PCR. Notedly, the experimental setup varies compared to the native PCR variant.
The present article covers information on multiplex PCR, its protocol, procedure and primer preparation. In addition, I will also discuss the advantages, applications and shortcomings of the present technique.
This article will add more value to your Polymerase chain reaction knowledge and helps you to achieve more success in PCR technology.
Stay tuned,
Key Topics:
What is Multiplex PCR?
In 1988, Jeffrey S Chamberlain and coworkers developed the concept of multiplex PCR. They had amplified different dystrophin gene locus from muscular dystrophy patients using a single reaction setup.
Put simply,
The multiplex PCR often abbreviated as mPCR is an excellent variant of the conventional PCR in which using different sets of primers, different DNA templates or various regions of a gene can be amplified, in a single reaction.
It’s a combination of many separate PCR reactions, however, we need an excellent reagent cocktail to do this. Notedly, the reaction contains general PCR reagents such as dNTPs, reaction buffer, nuclease-free water, and Taq DNA polymerase.
Read about each individual component:
If it is a qPCR multiplex reaction, then we need additional reagents like oligo (dT) primers, TaqMan probe or SYBR green dye and reaction buffer.
The best benefit of using multiplexing is saving time. We need more reagents to achieve the amplification of each template, though.
Principle of Multiplex PCR:
Unlike conventional PCR settings, here, many separate reactions, a machine can complete in a single tube.
Two common techniques for that are;
- Multi-template multiplex PCR
- Uni-template multiplex PCR
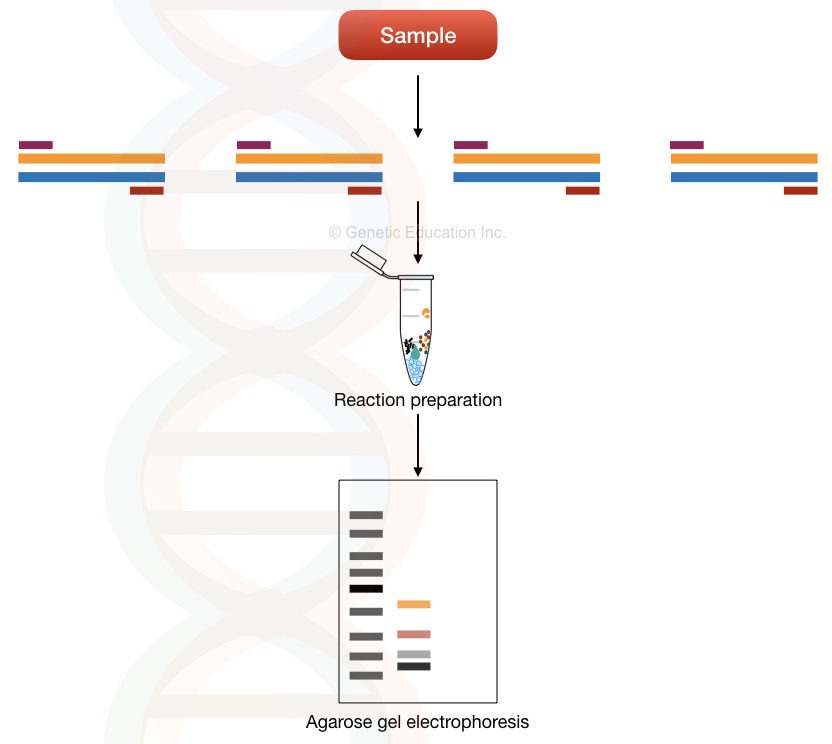
The multi-template multiplex PCR principle relies on the amplification of different templates in a single reaction. Meaning, it amplifies various templates present in a single sample using different primer sets.
Each set of primers is unique to each template. And therefore can amplify the target/complementary sequence (template) only. See the image below, it makes things more clear.
Multi-template multiplex PCR is used in microbial genetics, pathogen identification and detection of microbes. Here, it quantifies, amplifies and identifies microbes/pathogens present in a sample.
The present technique can’t be useful in detecting single-gene disorders.
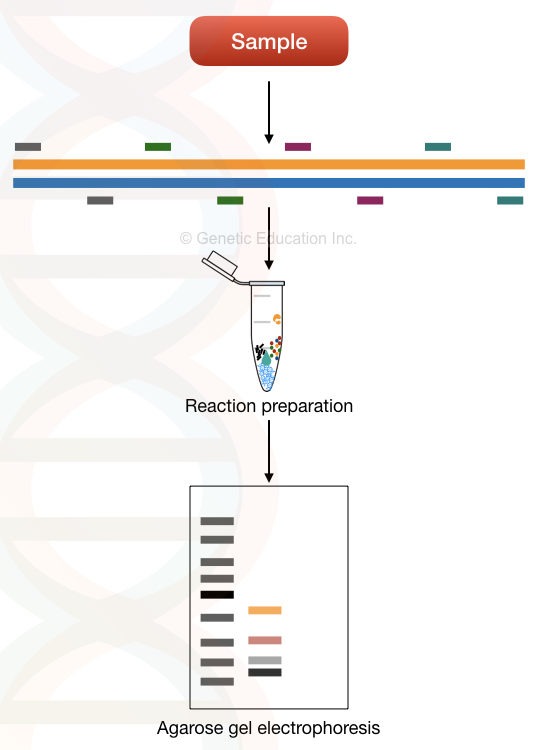
On the other hand, the uni-template multiplex PCR uses various loci of a single template DNA to amplify. Meaning, it amplifies many regions of a single gene or template using a different set of primers.
For example, in order to detect 5 different mutations of the beta-thalassemia, we have to prepare a single reaction for a single beta-globin gene.
5 different sets of primers, amplify 5 different mutation loci on a single beta-globin gene. That’s how things work in uni-template multiplexing.
It has tremendous applications in screening single-gene inherited disorders. Notedly, the first multiplexing reaction was performed on various loci of the single dystrophin gene.
It is widely applicable in detecting and genotyping loci. The image below explained the whole process,
Factors affecting multiplex PCR:
Common factors that influence a PCR reaction are many but two important factors that decide the fate of the multiplex reaction are,
- Primer designing
- Reagents utilized
Primer designing:
Primer design has a very critical role in multiplex PCR success as wrong primers may negatively influence the reaction. An inappropriate primer set either slips amplification of the target, can’t amplify anything or goes non-specific.
It means, only the correct primer sets may amplify loci. Primer length, melting temperature, GC content, hairpin structure and dimer formation capacity should be taken into consideration while designing primers.
Remember, every primer set used in a single multiplex reaction must be unique and don’t even cross-react with each other. Plenty of software is now available to design primers for multiplex PCR.
If you wish to design primers for multiplex PCR you can explore this tool available on PREMIER Biosoft. Use it: PrimerPex.
If you wish to read about primers and designing, read our article on this topic: PCR primer design guidelines.
Multiplex PCR reagents:
The quality and quantity of each reagent have a significant impact on multiplexing. For instance, higher concentration leads to non-specific binding while lower concentration fails to amplify the template, thoroughly
If you use a very low concentration of let’s say some reagent, it even can’t amplify the target loci. Every reagent has decent importance in the reaction and is consequently applied as per the requirement or provided by the manufacturer.
A few common reagents, a multiplex reaction needs are enlisted here:
- Taq DNA polymerase
- dNTP cocktail
- PCR reaction buffer
- Sets of primers
- Template DNA
- Nuclease-free water
- A TaqMan probe or SYBR green dye
- Multiplexing buffer
| Reagent | Utility |
| Taq DNA polymerase | An enzyme that amplifies the DNA |
| dNTPs cocktail | Provides individual dNTP during the synthesis. |
| PCR reaction buffer | Increase the efficiency of the reaction and decrease non-specific binding |
| Sets of primer | Do amplification by annealing with the template. Provides a free 3’ end to the polymerase. |
| Template DNA | A target DNA that has to be amplified |
| Nuclease-free water | Gives volume to the reaction and dilutes reagents. |
| A TaqMan probe or SYBR green dye | Used for quantification |
| Multiplexing buffer | Increase reaction efficiency, and specificity and reduce chances of non-specific binding. |
Multiplex PCR procedure:
The PCR procedure initiates with DNA extraction. Using a DNA extraction kit or protocol, high-quality DNA is extracted, purified and diluted.
To prepare the reaction, all the reagents are first taken and thawed. Prepare a multiplex reaction as given below,
- First, add the master mix or dNTPs cocktail to the reaction tube.
- Add reaction or multiplex buffer to that. Usually, the reaction buffer may have ingredients such as MgCl2, DMSO, salt, or KCl.
- Add sets or primers simultaneously to the reaction.
- Add Probe or dye, if the assay is quantitative.
- Add the template DNA to the reaction.
- Make up the final reaction by adding nuclease-free water to each tube.
- After completing the reaction setup, immediately set up the thermocycling condition. Note that the thermocycler must be switched on before preparing the reaction.
- Setup reaction as per the manufacturer’s protocol or use your own standard protocol.
- Put all the tubes in the machine and close the lead.
- Now start preparing agarose gel, if the PCR is a conventional one.
Multiplex PCR protocol:
In order to make you understand how the multiplex reaction is prepared, I am explaining the reaction preparation in this section for 4 different primer sets. Assume that it’s a conventional quadruplex PCR reaction.
- Dilate the extracted DNA and check the purity. Use 50 ng DNA in the reaction.
- Add dNTP mix up to 13 µL in the reaction with 10 mM contention.
- Use a mastermix buffer or reaction buffer 1X, if given in 10X. Use up to 7 µL.
- Add (10 pM) each set of forward and reverse primer sets, 1 µL each. Meaning, 4 for forward and 4 for reverse primers (4 *4).
- Add (50ng) 5µL template DNA to the reaction.
- Make up the final volume by adding 4 to 5 µL of nuclease-free water.
| Component | Final Concentration | Quantity |
| Master mix | 1X | 13µL |
| PCR reaction buffer | 1X | If needed |
| Forward primer (four sets of primers) | 10pM | 1 x 4= 4µL |
| Reverse primer (four sets of primers) | 10pM | 1 x 4= 4µL |
| Template DNA | 50ng | 5µL |
| Water | 4µL | |
| Total | ——————————- | 30µL |
After preparing the reaction do PCR setup as according,
| PCR Steps | Initial Denaturation | Denaturation | Annealing | Extension | Final extension |
| Temperature | 90 ̊C-95 ̊C | 90 ̊C-95 ̊C | 55 ̊C-6o ̊C | 72 ̊C | 72 ̊C |
| Time | 5min | 1min | 50sec | 1min | 7 min |
| ——————– | ——————- | 25-28 cycles | ————— | ——————— |
Advantages of multiplex PCR:
- Firstly, it can handle the amplification of various templates in a single reaction or tube.
- The technique is rapid, time-saving and utilizes less manpower.
- It is cost-effective. Meaning, multiplexing saves reagent, time and power hence it is cheaper.
- Each amplicon works as an ‘internal control’ for another reaction and therefore helps to encounter false-positive results.
- It can predict the quality of the template DNA by comparing different amplicons of the same gene or DNA.
- It gives more information by using less starting material (template).
- Technically, the assay needs less consumables, chemicals and other utilities.
- In addition, it reduces the chances of pipetting errors.
Limitations:
Though the present technique has more advantages than any other PCR variant, it has some serious shortcomings.
- First, the chances of non-specific bindings, as well as un-amplifications, are too high. Meaning, it has a very high reaction failure possibility if not performed well.
- It can’t amplify longer templates effectively. The chances of amplifying every primer set using a longer gene template are fewer. Usually, up to 100bp gene template, it can amplify precisely.
- We can’t multiplex all types of templates. It is only applicable to a few types of templates. Shorter templates do well.
- In addition, the correct primer set can’t be prepared every time. Meaning, every primer set isn’t compatible with each other.
- Increasing the number of templates to amplify needs more primer sets and reduces the chances of success. Hence, the more the templates, the more the chances of reaction failure.
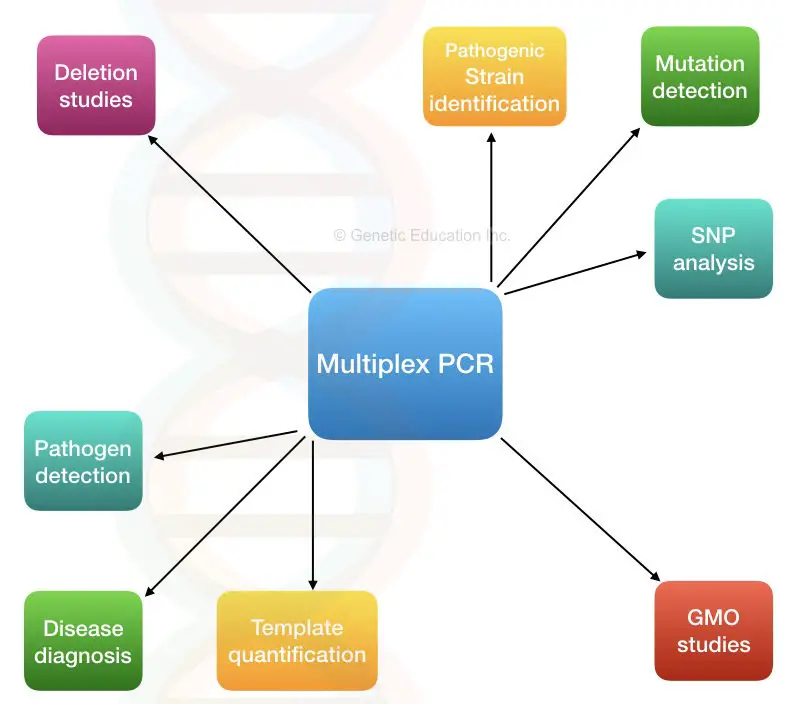
Applications of multiplex PCR:
Multiplexing reactions make the task easy to perform and quickly so applicable in many scientific fields.
Microbial genetics- Pathogen detection, identification and characterization:
Traditional microbiology techniques have time-consuming, contamination-prone, tedious culturing steps. PCR replaced it with its higher accuracy, sensitivity and time-saving procedure.
Various strains of pathogens, and different pathogens present in a sample can be identified and studied using the multiplex PCR approach. Researchers first isolate DNA and amplify it using various primer sets.
Computational tools help in the identification and characterization of microbes present. Read more on the present topic: Importance of microbial genetics.
Metagenomic studies:
Metagenomic studies highly rely on the multiplex PCR assay. DNA obtained from any environmental sample such as soil, water, dead leaves or biomass has been processed using this approach.
The machine amplifies each microbial DNA present in the sample using each set of respected primers. Computational tools evaluate results.
Detecting infection:
One of the most important applications is in detecting various infective microbes present in a sample. Various strains of SARS-CoV have been detected using this approach in recent days.
Ready-to-use kits for VZV, CMV, T. Gondii, influenza and adenovirus are now available for multiplex PCR.
SNP genotyping:
A single conventional PCR reaction can encounter a single SNP and consequently identifies only a couple of genotypes. Whereas using the multiplex PCR approach many SNPs can be ruled out in a single reaction.
This makes it possible to study many genotypes at once. However, it has a limited range of multiplexing. Including more primer sets for more SNPs elevates the chances of reaction failure or false-positive reaction.
In the present article, we have explained the process of genotyping: What is a genotype?
Studying GMO:
Scientists prepare Genetically Modified Organisms by altering their genomic portion. It is possible to amplify and study many genes of interest in a single reaction.
Once a gene is introduced, PCR helps to validate many templates in a single reaction.
Mutation detection system and polymorphism study:
Yet another important application is in detecting various mutations and studying genetic polymorphism in species. The best example is the DMD gene. Multiplex PCR amplifies different exons of the DMD gene.
Results can be studied on an agarose gel or computationally. As we said, it can study allelic variations too.
Qualitative and quantitative analysis of template:
For critical experiments such as sequencing and microarray, the quality and quantity of the template DNA matter a lot! A good quality template elevates the chance of getting results.
This technique can quantify the number of templates present in the sample as well as assess the quality of the template. Many temperate ‘regions’ (DNA loci) can be studied in a single qPCR multiplex reaction.
Besides, other applications are in linkage analysis, forensic analysis, gene transfer study.
Fact: Multiplex PCR can find 98% of deletions in the DMD gene among all exons.
Optimizing the multiplex PCR:
Designing primer, adding the accurate amount of reagent and setting correct thermocycling conditions isn’t only sufficient to get results. We need to optimize our multiplex reaction in order to get good amplification.
For instance, you can use PCR additives such as MgCl2, KCl, DMSO or albumin in the reaction, additionally.
Developing various multiplexing PCR assays needs expertise, experience in PCR, huge research, trial and error experiments and high-end instruments.
Besides, assay success depends on some common things, for example, as we are amplifying many templates here, we need each reagent a bit more, that’s obvious.
More units of Taq DNA polymerase, more PCR buffer, a higher concentration of dNTPs and primer sets should be advisable.
As we said, primer designing has a pivotal role here. This should be done precisely or by experts only.
Keep in mind that the primers shouldn’t be complementary to each other, otherwise, they bind to one another and fail the reaction. Choose the primers having less primer-dimer forming capacity.
The GC content of each primer must be between 45% to 60%.
The advisable range of melting temperature should be between 55°C to 60°C.
To achieve even amplification for all templates, the annealing temperature of each primer set should be nearer enough, ideally. Primers that have lengths up to only 30 nucleotides are tolerable.
Primers between the length of 20 to 25 nucleotides are ideally recommended.
Besides, yet another crucial assay factor is the ‘PCR cycles’. More cycles cause reaction failure or truncated amplification as nearly no reagents are available at later cycles.
On the other side, fewer PCR cycles abort amplification prematurely. Consequently, all the loci aren’t amplified properly. Correct PCR cycling conditions should help achieve amplification in multiplex PCR.
Ideally, 25 to 30 cycles are sufficient, though micro-standardization must be required to achieve good results.
The critical parameter that must be satisfied is the pattern of amplification. Design a multiplex PCR reaction in such a way that gives a well-separated, distinguishable banding pattern.
If primers amplify regions too nearer, the differential banding pattern can’t be seen.
In summary, making only a PCR reaction isn’t sufficient for multiplex PCR, we should have enough knowledge to perform it. That’s why it needs experience and expertise.
How to perform a multiplex qPCR test?
The process of multiplex qPCR test is similar to the conventional multiplex PCR, and needs a few extra reagents. However, it quantifies various templates present in a sample and therefore has great utility in infectivity and diagnostic studies.
Primer designing, assay planning and primer ordering steps are completed earlier before doing the assay.
First, like other PCR reactions, we need a pure DNA sample extracted using a DNA extraction kit.
All the pre-reparation steps such as cleaning the PCR bench, collecting and thawing reagents and preparing SOP have been completed first.
Prepare a qPCR reaction using the table below,
| Component | Quantity in reaction | Final concentration |
| High fidelity reaction buffer with MgCl2 | 2 μL | 1X |
| dNTP mix | 2 μL | 200 μM |
| Template DNA | 3 to 5 μL | 100 ng |
| Forward primer mix | 3 μL | 30 ng |
| Reverse primer list | 3 μL | 30 ng |
| High fidelity DNA polymerase | 0.5 μL | 2.5 U |
| TaqMan Probe | As per the manufacturers’ advice | |
| Nuclease-free water | 3 to 5 μL | |
| Total | 20 μL |
Once the test preparation completes, place tubes in a machine and run the PCR. The result of the test is evaluated, as explained in the above section.
During the 202 Pandemic, the CDC- center for disease control have announced an RT-PCR multiplexing assay for the SARS-CoV-2 coronavirus. As of 2020 or early 2021, many variants of the coronavirus have ruled the world badly.
Some of them are alpha, delta, epsilon and UK variants of SARS-CoV-2. A multiplexing assay can detect many strains present in a single sample. Recently 90 years old women detected positive for two different coronavirus strains, which were processed through multiple RT-PCR.
In summary, the technique saves time and cuts the cost of detecting SARS CoV-2.
Conclusion:
In comparison with conventional PCR, multiplex PCR has more advantages than shortcomings. It literally helps us in many ways and therefore scientists use it.
However, as the assay has less specificity, it isn’t advisable for critical experimentations. You need to develop your own multiplexing assay by optimizing temperature, cycling conditions and reagents which suit your reaction.
As I said, multiplex PCR isn’t possible for all types of templates, do throughout research before planning to develop an assay. Though many ready-to-use kits are now available, multiplexing reactions have been less used in diagnosis.
Sources:
- Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000;13(4):559-570. doi:10.1128/CMR.13.4.559.
- Oscorbin IP, Shevelev GY, Pronyaeva KA, et al. Detection of SARS-CoV-2 RNA by a Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Melting Curves Analysis. Int J Mol Sci. 2021;22(11):5743. Published 2021 May 27. doi:10.3390/ijms22115743.
- Markoulatos P, Siafakas N, Moncany M. Multiplex polymerase chain reaction: a practical approach. J Clin Lab Anal. 2002;16(1):47-51. doi:10.1002/jcla.2058


