“The Polymerase chain reaction is an in vitro DNA synthesis method in which DNA is amplified using the Taq DNA polymerase enzyme.”
Related article: 10 Strategies to Achieve Excellent PCR Amplification.
The polymerase chain reaction is a common technique practiced in genetic laboratories because it is a basic requirement for a genetic or molecular lab.
Replication is a process of DNA synthesis, however, for us mimicking replication in a lab isn’t possible. But after the discovery of the thermostable DNA polymerase, the dream of synthesizing DNA in a lab has come true. What is the problem with the normal DNA polymerase? well, it can not work at a higher temperature.
A thermostable Taq DNA polymerase, isolated from the hot water bacteria can synthesize DNA even at a higher temperature. Using ingredients such as dNTPs and other PCR enhancers along with Taq, one can synthesize DNA in PCR.
The Polymerase chain reaction is one of the emerging scientific techniques and has infinite opportunities in research as well as diagnostics. Different variations in the native PCR helps in the development of different techniques for different applications.
Allelic-specific PCR, Real-time PCR, reverse transcriptase PCR, Hot start PCR, and nested PCR are some of the common PCR types used in every genetics lab so often.
In the present article, we will understand the PCR- polymerase chain reaction, starting from basics to advance. Furthermore, we will also discuss some of the important types of PCR used to enhance PCR results.
Overview of the article,
- History and overview
- Definition
- Requirements
- Principle of PCR
- Steps in PCR
- PCR machine
- PCR reagents
- PCR procedure/ protocol
- PCR results
- Applications of PCR
- limitations of PCR
Key Topics:
History and overview:
In 1983, Kary Mullis described the technique of in vitro gene amplification and named it as a polymerase chain reaction. Later on, he was awarded the Nobel Prize for his finding.
However, the story of PCR was begun when the Taq DNA polymerase was isolated from the thermostable bacteria. In 1996, Thomas D Brook had discovered the bacteria from the hot spring of water and named it Thermus aquaticus. Later on, in 1976, Chien et al., isolated DNA polymerase from Thermus aquaticus named it as Taq DNA polymerase.
The overall idea of the Polymerase Chain Reaction is to get copies of a DNA or gene we wish to study. we can’t visualize a few DNAs that is why we need to amplify DNA. After the isolation of thermostable Taq DNA polymerase, the idea of temperature-dependent amplification came into the picture.
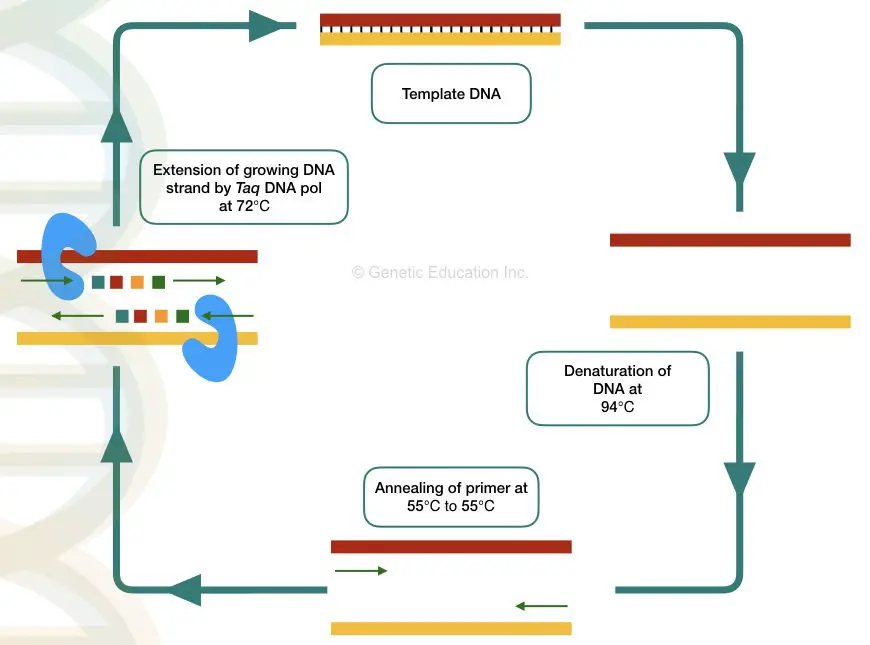
The PCR- polymerase chain reaction is a temperature-dependent process of DNA amplification. The machine used in the PCR technique is known as a Thermocycler. Let’s understand each terminology properly.
The word PCR is made up of Polymerase– Taq DNA polymerase + chain– cyclic reaction + reaction– biological activity. Taq DNA polymerase governed cyclic reaction is known as PCR.
Thermocycler: The machine thermocycler provides various temperatures for each step to complete. Denaturation, annealing, and extension of DNA occur at different temperatures thus the machine is known as a thermocycler. The overview of every PCR cycle at various temperatures is given below,
PCR definition:
“A common genetic tool- a laboratory technique used to obtain multiple copies of target DNA fragments using Taq DNA polymerase in a temperature-dependent reaction is called a PCR- a polymerase chain reaction.”
Theoretically, the definition of the PCR can be as stated,
“PCR is a technique in which using the dNTPs, primers, Taq DNA polymerase, and template DNA, artificial gene synthesis can be done.”
Or we can say,
“PCR- a polymerase chain reaction is a cyclic temperature-dependent reaction used to amplify the gene of interest.”
In short, we can define PCR as,
“An in vitro DNA amplification or synthesis technique is known as PCR.”
PCR requirements:
Chemicals: dNTPs, distill water, PCR reaction buffer, enzyme Taq DNA polymerase, primers, and template DNA.
Instruments: thermocycler, spinner and agarose gel electrophoresis unit.
Other utilities: PCR tubes, stands, pipettes, tips.
Principle of PCR:
The principle of the PCR is based on the temperature variations of heating and cooling- thermocycling reaction divided into three steps:
Denaturation- The dsDNA becomes single-stranded at a higher temperature during denaturation. Here hydrogen bonds between two DNA strands break.
Annealing- The primer binds or anneals to its exact complementary sequence on a DNA during the annealing step. The primer provides a site for the initiation of synthesis.
Extension- Taq DNA polymerase uses the 3’ end of the primer and starts DNA synthesis by adding nucleotides to the growing DNA strand.
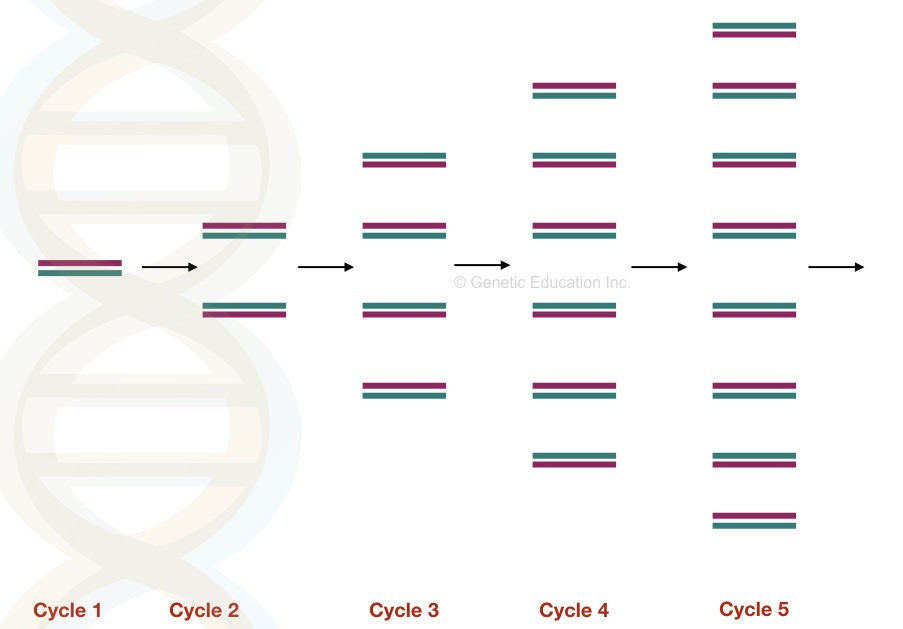
All three steps are repeated for 25 to 40 cycles and in each cycle the DNA becomes double.
Note: Amplification is a DNA copying process just like DNA replication.
Do you know?
The PCR machine was not always an automated machine.
The first PCR machine was a series of three different water baths with three different temperatures. The traditional machine did not have a digital display or a temperature controller. In those days, scientists have to transfer PCR tubes in each water bath manually at least 35 times.
Each water bath had a thermometer for monitoring temperature. Karry Mullis had achieved PCR amplification through this process. However, in Year 1985, PerkinElmer introduced the first automated PCR machine. Because of that PerkinElmer is one of the pioneers and tech giant companies in making PCRs.

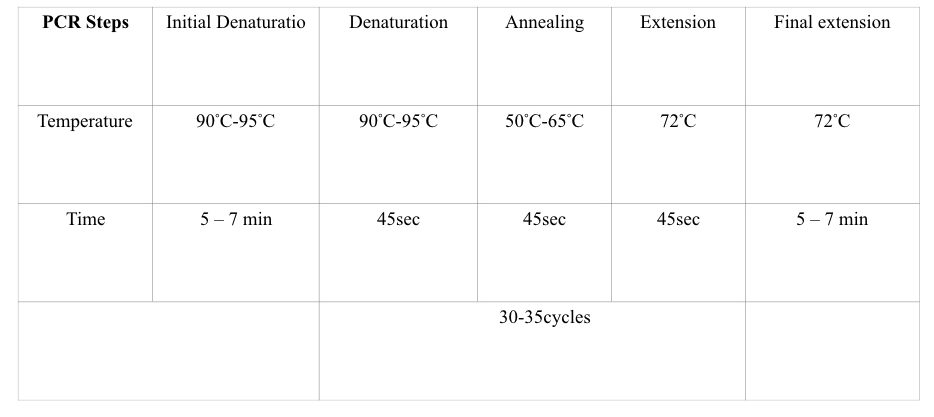
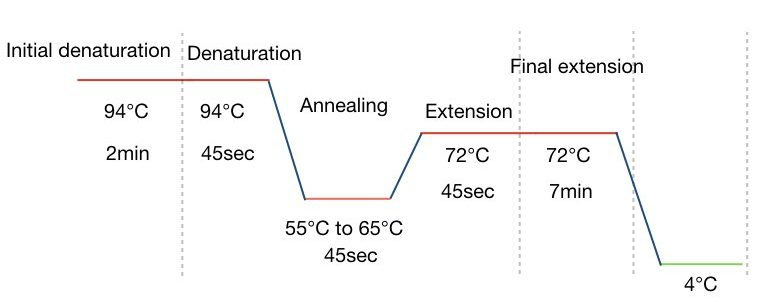
PCR steps:
Various temperature zone governs each PCR step, viz denaturation, annealing, and extension followed by a single initial denaturation and final extension steps. In each step, different reactions occur.
PCR step 1: Denaturation
- Temperature: 90°C to 95°C
- Time 30 sec to 90 sec
In a denaturation two single-stranded DNA form from the double-stranded one. At 94ºC temperature, the double-stranded DNA opens up by breaking hydrogen bonds. The process of denaturation is followed by the initial denaturation for 5 to 7 minutes at the same temperature.
PCR step 2: annealing
- Temperature: 55°C to 65°C
- Time: 30 to 60 sec
After the denaturation, primer anneals to ssDNA at its exact annealing temperature. Based on the GC content of primers, every primer has its own annealing temperature.
The annealing temperature is usually ranging from 55ºC to 65ºC. Annealing temperature lower than that leads to non-specific bindings while higher temperature leads to amplification failure. 45 seconds to 1 minute are enough for the second step, annealing for more than 1 minute causes non-specific amplification.
Related article: What is Annealing Temperature in PCR? How to Calculate and Set It?
PCR step 3: extension:
- Temperature: 70°C to 72°C
- TIme: 45 Sec
After the binding of the primer, it’s time to expand the DNA strand. Here in the extension step the Taq DNA polymerase comes into action and adds dNTPs to the DNA strand. The temperature for the extension is 72ºC for 45 seconds.
After completing all steps one more time the final extension is performed for 7 minutes. The graphical representation of each PCR step is explained in the figure below:
Time-duration for PCR:
- 1 hour to 4 hours
PCR reagents:
Template DNA, PCR primers, dNTPs, Taq DNA polymerase and PCR buffer are the major reagents of PCR reaction. The composition and quantity of each reagent are very important. A single μL variation in any of the reagents leads to reaction failure.
Template DNA:
The template must be DNA only. Plasmid DNA, bacterial DNA, cDNA, or gDNA can be utilized as a template. The template DNA should be highly purified DNA that has a purity of around ~1.80 and a quantity of up to 200ng. The DNA works as a substrate for an enzyme when it is denatured.
The good quality of extracted DNA can boost the resulting efficiency of the polymerase chain reaction. The ideal concentration of gDNA for PCR reaction is 30ng with a 260/280 absorbance ratio of ~1.80.
Concentration: 30 to 50ng.
PCR Primer:
Another important PCR ingredient is PCR primers. RNA primer governs the replication reaction, normally, however, DNA primers are utilized during PCR instead of RNA primers. The Taq DNA polymerase doesn’t have proof-reading activity thus it can’t remove RNA primers. Probably, this is the reason, we are using DNA primers in PCR.
(The RNA primer is replaced during the proofreading activity in the replication which is not possible in the case of Taq DNA Polymerase). For more detail on DNA replication please read the article: DNA Replication class 1: General process of DNA replication
(The RNA primer is replaced during the proofreading activity in the replication which is not possible in the case of Taq DNA Polymerase).
The PCR primers are artificially synthesized oligonucleotide sequences of DNA ranging from 18 to 22 bases in length, short DNA sequences which anneal at the single-stranded template DNA at its exact complementary position.
For increasing the efficiency of primers, we should follow proper guidelines while creating primers. GC content, melting temperature, length, and primer-complementation capacity of primers are key factors for primer designing. For more detail on the primer design guide, read the article: PCR primer design guidelines.
Generally, 10pmol of each primer is sufficient for a PCR reaction.
Concentration: 10 to 12pMol.
dNTPs:
Deoxynucleotide triphosphates are artificially synthesized nucleotides that bind to the growing DNA strand. With the help of the Taq DNA polymerase, the dATP, dGTP, dCTP and dTTP bind at their complementary nucleotides on the growing DNA strand.
1mM to 2mM of each dNTPs are sufficient for 25μL of PCR reaction, For more detail on how to prepare a working dNTP solution, read the article: The Function of dNTPs in PCR reaction
Concentration: 200-250μM each.
Taq DNA polymerase:
The PCR technique is entirely based on the activity of Taq DNA polymerase. If Taq DNA polymerase was not discovered, the PCR might not be discovered.
Thermostability, the unique property of the Taq makes amplification possible during PCR. Thermostability means it can work finely at a higher temperature. Notably, no other bodily enzyme can function at a higher temperature of more than 37ºC.
The Taq DNA polymerase settles at the ssDNA- primer junction and utilizes it as a substrate for the catalytic reaction. In the final step of extension, using the substrate it starts dNTP insertion.
1 unit of Taq is sufficient for a 25μL PCR reaction. For more detail on Taq DNA polymerase read the article: Function of Taq DNA polymerase in PCR.
Concentration: 1 to1.5 unit.
PCR buffer:
PCR enhancers help to boost reaction and amplification efficiency thus PCR buffer is as important as other ingredients. Additionally, the PCR buffer maintains the constant pH of the reaction nearly 7.9 to 8.5 by keeping the constant chemical environment for the PCR reaction.
The pH of the buffer is controlled by the addition of Tris.
Mgcl2, DMSO, KCl, albumin, betaine, BSA, glycerol, (NH4)2SO4, and formamide are some of the chemicals commonly used in the PCR buffer. The composition of each ingredient may vary from manufacturer to manufacturer.
However, in each PCR buffer, the MgCl2 must be included because it is worked as a cofactor for the Taq DNA polymerase. For more detail on PCR buffer ingredients read the articles:
Concentration: 1X or as per requirement.
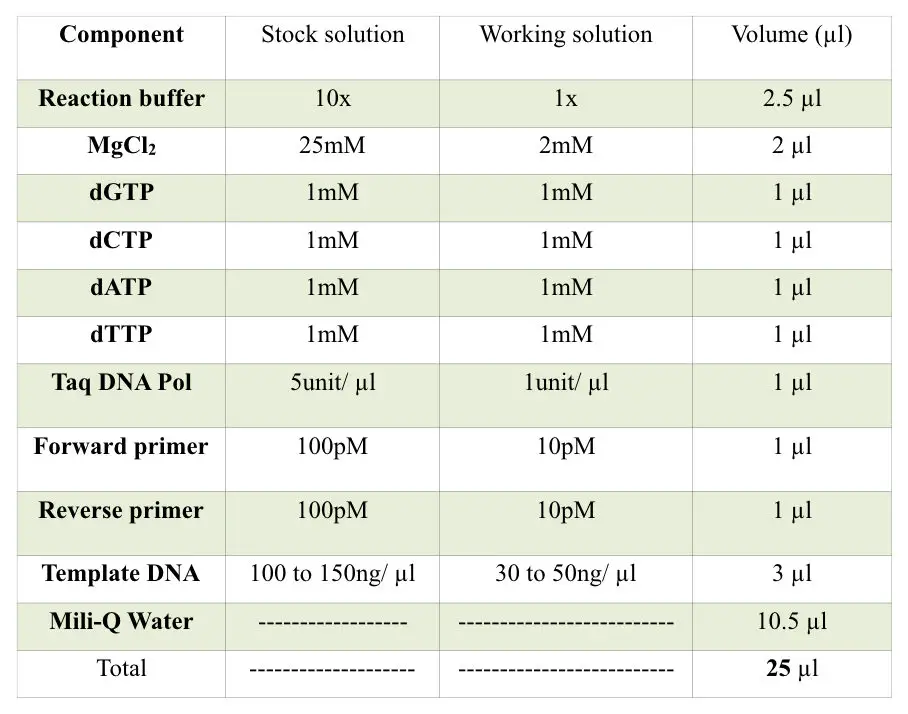
The PCR machine
The PCR machine is known as a thermocycler. This machine is simply a heating block (just like our iron) which provides the constant temperature and even rapidly changes between two temperature states.
The machine has a lower block of metal having deep wells for putting PCR tubes. Also, the temperature of the inner environment is maintained by the heating block present on the upper side of the lead.

Further, the machine contains the display, power on and off switch, and cooling assembly. The machine has the ability to heat and cool the PCR tube in a short period of time.
PCR procedure/ protocol:
Pre-preparation:
For any molecular genetic experiment, pre-preparation plays an important role in getting good results.
- Before starting the reaction, one must have to be ready for doing the lab work, for that, wear a lab coat, gloves, a mouth cap, and a head cap.
- Clean the PCR reaction preparation area and arrange all other utilities nearby the reaction preparation.
- Now take reagents from the deep freeze and thaw all the reagents properly.
Reaction preparation:
Take a sterile PCR tube and start adding reagents as shown in the table.
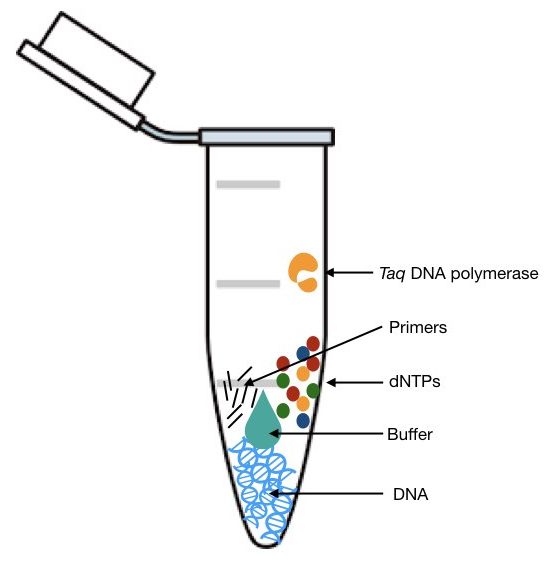
starts adding reagents in a sequential manner to reduce the chance of error.
If you have a ready-to-use mastermix, you can add it directly, this will save time and increases the efficiency of the reaction.
After the completion of reaction preparation, close all the tube caps and spin it properly, so that all the reagents mix well.
Now put the tubes in the PCR machine one by one in the pre-set PCR protocol. Remember: don’t waste time setting protocol during the PCR, set it before the reaction preparation, and immediately run the PCR.
Meanwhile start preparing the gel for agarose gel electrophoresis, because it will also take time for around 60 to 90 minutes.
Key to success in PCR
I suggest you to measure every reagent and ingredient properly to get uniform and accurate results.
Post-preparation:
- After completion of the PCR reaction, turn off the machine and collect all the tubes in an “orderly manner”.
- Rest tubes for some time in a freeze before doing agarose gel electrophoresis.
- You can also rest it for the next day, no problem with it.
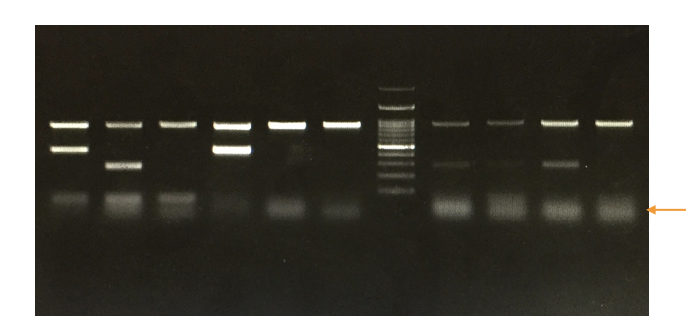
PCR results:
We have covered an amazing article on analyzing and interpreting agarose gel electrophoresis results, that portion will master you on this.
Anyway, we will explain to you how to interpret the results of PCR in brief,
Run a DNA ladder along with the PCR amplicons so that we can analyze the results.
Based on the migration of DNA fragments in the gel and our in silico PCR or primer 3 results we can assume what size our PCR amplicons are.
See the image below,
Application of PCR
We have covered an amazing in-depth article on applications of PCR, you can read it here: 50 Powerful applications of PCR.
Here I am only enlisting some of the important applications of PCR.
The PCR has numerous applications in biological research as well as diagnostics.
Diagnosis of inherited disease: PCR is most routinely used in the diagnosis of some inherited diseases such as sickle cell anemia, thalassemia, MTHFR gene mutation, etc. This technique is appropriate for single-gene disorders. The result is 99% accurate as compared with other methods.
Microbial identification: the microbial culture technique is traditional and time-consuming and the chance of infection is also high in the case of culturing. In modern days, PCR is used in the identification of microbes. The bacteria’s unique DNA sequence is targeted for the identification of particular bacteria. It will give a result within 3 to 4 hours.
Additionally, PCR is also applicable to the diagnosis of infectious diseases such as HIV or HPV. Again the method is the same as the identification of microbes. The unique DNA sequence of a particular virus is targeted for identification. This will give a result within an hour.
PCR is used in the identification of genetic carriers as well. The heterozygous condition of the disease can be easily identified using PCR amplification.
DNA fingerprinting and genetic imprinting: PCR is the first choice for DNA fingerprinting. For more detail on DNA fingerprinting read the article: DNA fingerprinting
Criminal verification, identification of a person, and material cell contamination can be detected using DNA fingerprinting.
The PCR is one of the best techniques for marker assistant selection. RFLP, AFP, RAPD, STS, VNTR, and STR are some of the PCR-based techniques.
PCR is applicable in the prenatal diagnosis of inherited disease as well.
PCR helps in detecting cancer genes and infections.
Further PCR is applicable to sex determination and sex identification.
Apart from mutation detection, PCR is useful in gene expression studies too. The expression of a particular gene can be measured using RT PCR. It is even applicable in gene cloning.
mRNA studies are also possible due to the reverse transcriptase PCR and we can calculate gene expression through it.
PCR amplification is one of the important steps in DNA sequencing and microarray.
The PCR is also useful in the validation of personalized medicines.
The PCR is used in;
- Gene editing
- Gene manipulation
- Genetic engineering
- RNAi research
- DNA and RNA quantification
- cDNA and gDNA library preparation
- Developing new assays
Limitations of PCR
Novel mutations can not be found using PCR, we have to do sequencing for that.
Also, Multigenic disorders cannot be detected using PCR.
We can not identify structural and numerical chromosomal anomalies through PCR.
Types of PCR:
Conventional PCR:
The simplest version or the original PCR technique utilizes only a simple Taq DNA polymerase and no modifications called conventional PCR.
Gradient PCR:
Gradient PCR is one of the widely used modifications of native PCR in which for optimizing the PCR reaction, different temperature gradients are created in a machine.
Using these different temperature gradients, the template DNA amplification efficiency can be checked. The best annealing temperature can be selected for further consecutive reactions.
Besides this, the efficiency of different PCR enhancers can also be checked at different temperatures using gradient PCR.
Read more: Gradient PCR
Hotstart PCR:
The benefit of using the Taq DNA polymerase in the PCR reaction is its stability at a higher temperature, however, it is also its limitations.
As it is also able to synthesize the DNA at a lower temperature too, using the hot start modifications, the Taq is inserted only at the time of denaturation.
For doing that, different strategies of inactivating Taq DNA polymerase early in the reaction are available. One of them is the use of enzyme liked antibody.
At a higher temperature, the antibody released the enzyme in the reaction. However, the main objective of the hot start is to activate Taq only when the reaction starts.
Read more: Hot start PCR
Realtime RT PCR:
Yet, another amazing modification of the native PCR is the real-time PCR in which using the fluorochrome chemistry, the template DNA can be estimated.
A probe attached with the fluorochrome emits fluorescence once it is hydrolyzed from the template and the template is measured.
The amount of fluorescence emitted is directly proportional to the amount of DNA present in the sample.
This is the principle of real-time PCR which is now widely used in diagnostic and microbial identification.
Here the catch is the use of the colored molecule, although, different types of probes are used for different applications.
Read more: Real-time PCR
Reverse transcription PCR:
Reverse transcription PCR is actually a variant of real-time PCR in which instead of DNA the amount of RNA can be measured.
An enzyme called reverse transcriptase converts the total mRNA into the cDNA which is measured using the same chemistry of the real-time PCR.
Thus the amount of the mRNA present in a sample can be estimated using this type of PCR.
As we know, the total mRNA translates into protein, therefore the gene expression can be measured using reverse transcription PCR.
Read more: Reverse transcription PCR
Multiplex PCR:
One of the major limitations of the PCR reaction is that only a single template can be amplified in a single reaction.
Multiplex PCR is a modification using which multiple templates can be amplified using a single set of primers or a single template can be amplified using the multiple sets of primers.
Multiplex PCR is widely applied in the real-time PCR assay for quantification of multiple templates or screening of multiple mutations in a single assay.
Though the method is similar, optimization must be required for developing different multiplex protocols.
Read more: multiplex PCR
ARMS PCR:
The amplified refractory mutation system is a unique type of PCR reaction set up in which different alleles of the same gene can be amplified using ARMS PCR and therefore it is also called an allelic PCR.
Generally, two pairs of primers- one for wild type allele and one for a mutant allele are used to amplify two different alleles.
A primer set for the wild-type allele can not amplify the mutant allele and thus a single DNA band for the homozygous allele is obtained.
A primer set for mutant type allele can not amplify the wild type allele and thus a single DNA band for a mutant homozygous is obtained.
However, two different DNA bands one for wild type allele and one for mutant type allele is obtained in heterozygous DNA.
Read more: ARMS or allele-specific PCR
Touch down PCR:
Now, this modification is my favorite one! Because it always gives positive results in all assays.
In the touchdown PCR, by gradually decreasing the annealing temperature, the specificity in a PCR reaction can be increased.
First, set up the annealing temperature 10ºC above the real annealing temperature and then set up each PCR cycle with a decrease in temperature 1C per each cycle until 50ºC or 55ºC.
The main objective of doing this is to increase the specificity of the PCR amplification without compromising the specificity.
Read more: Touchdown PCR
Nested PCR:
Using one of the nested PCR along with the flanking primers, the efficiency of the PCR reaction can be increased by employing the nested PCR methods.
Read more: nested PCR
Colony PCR:
A rapid, high throughput PCR method in which the insert or the plasmid DNA is amplified directly from the bacterial colony.
For that, the bacterial colonies are taken and PCR is performed directly on it which helps in amplifying insert directly without extracting plasmid DNA.
However, two sets of primers are used for that, one for plasmid specific and one for amplifying the rest of the DNA.
Read more: Colony PCR
In situ PCR:
In situ-PCR is yet another excellent method for rapid amplification of a sample DNA. in this method the amplification of target DNA is done directly on the side or in situ. All the reagents such as dNTPs, primers and PCR buffers are added directly on the slide to do PCR.
This method is widely used for paraffin-embedded tissues or for formalin-fixed tissues.
Read more: in situ PCR
Immuno PCR:
Immuno PCR is a combination of real-time PCR and ELISA methods.
Using the sensitivity of the ELISA method in the quantification, the specificity of the PCR reaction can be increased using the Immuno PCR.
Read more: Immuno PCR
In silico PCR:
The in silico PCR is a computational tool used to estimate or predict the results of actual PCR reaction.
We have covered an amazing article on a step-wise guide on how to do in silico PCR.
Read more: in silico PCR
Droplet PCR:
The droplet PCR is further, an amazing enhancement of the PCR quantification in which using the droplet; the amount of the template DNA is estimated
Droplet PCR is an assay used to estimate the amount of the template, especially, for sensitive assays such as quantification of pathogens.
Read more: Droplet PCR
Asymmetric PCR:
The Asymmetric PCR is used to amplify only a single DNA strand for DNA sequencing and probe hybridization. It needs more PCR cycles, a template-specific primers and annealing temperature.
Read more: Asymmetric PCR.
Conclusion
The polymerase chain reaction is a highly sensitive biological technique. The chance of cross-contamination is always high in the case of the PCR. Always perform PCR reactions in a sterile area otherwise the chance of the false-positive result will increase if any of the ingredients are contaminated.



Hello,
About your recip table , how Can you have 1mM of dNTP in the stock solution AND in the working solution ???
It shows that if you have 1mM of stock solution (which supplied by some of the companies) you have to directly use it as the working, no need to make a separate working solution.
Contrary, if your dNTP stock solution is 2mM, use half of it.