“Learn how and why scientists target a specific genomic region or gene(s) using targeted sequencing. How the technique works with some examples and applications in this comprehensive guide.”
What if you lost your key in your room? You have to search for possible places where it will be! Meaning, you have to do a ‘targeted’ search for your key. That’s what exactly geneticists do with targeted sequencing.
The ‘key’ here is a gene or sequence we wish to study and ‘places’ are the locations or regions in our genome. When we look for a specific gene(s), sequence(s) or chromosomal region, it is known as targeted sequencing.
Sequencing revolutions have strengthened our understanding of genetic traits and disease, however, the extremely talented whole genome sequencing, sometimes fails to give us what we want.
For example, if we are looking for rare variants, an unknown chromosomal region, a specific gene or alterations, WGS can not give us deep sequencing reads or accurate analysis for the region of our interest.
Targeted sequencing-like techniques help overcome these limitations and enable accurate and high-resolution analysis. So what exactly the targeted sequencing is? How does it work? And what are its applications?
Genetic Education Inc. focuses primarily on conceptual learning. Our team of experienced scientists is dedicated to re-discovering knowledge and presenting it understandably. We offer a sequencing master class for those interested in mastering the art of sequencing.
Stay tuned.
Important terminologies and their full name:
| Short name | Full Name |
| TS | Targeted sequencing |
| WGS | Whole-Genome Sequencing |
| Indels | Insertion and deletions |
| NGS | Next-Generation Sequencing |
| HLA | Human Leukocyte Antigen |
| BRCA | Breast Cancer Gene |
| CYP2D6 | Cytochrome P450 2D6 |
| PCR | Polymerase Chain Reaction |
Key Topics:
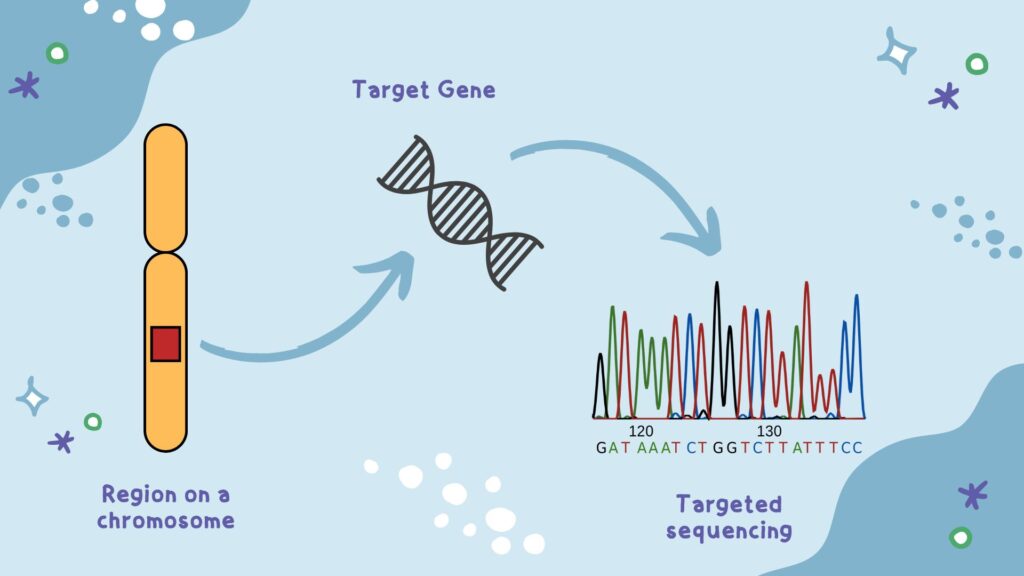
What is Targeted sequencing?
There are approximately 1650 mutations in BRCA1 and 1730 mutations in the BRCA2 gene, are reported to date (Munir et al./Nature). It’s impossible to study all these mutations using PCR amplification while whole genome sequencing can miss some of the rare variants from both genes.
This creates a loophole or critical error in screening or diagnosis of breast cancer. NGS-based targeted sequencing can effectively capture every mutation (common and rare) from the sample. That’s why TS is a crucial technique.
Targeted sequencing is a novel and focused sequencing approach that instead of sequencing the entire genome, focuses only on a dedicated subset of genes or genomic regions. This allows scientists to study specific genes, sets of genes, targeted genomic or chromosomal regions, associated alterations, etc.
TS makes it possible to perform an in-depth analysis of SNPs, indels and other alterations within a specific, targeted or rare gene. Thus, enables researchers to examine the variants or genes linked with a disease, condition or particular type of cancer.
You may wonder that the WGS can do this for the entire genome, and for many genes associated with disorders, then why do we need to target a specific gene or group of genes? To answer this question, we first need to understand the differences between WGS and TS.
Whole genome vs targeted sequencing:
WGS can sequence the entire genome of an organism while targeted sequencing can sequence only a specific region or gene from the genome. Thus, TS performs more accurate and precise analysis. Furthermore, it has higher resolution and sequencing depth compared to WGS.
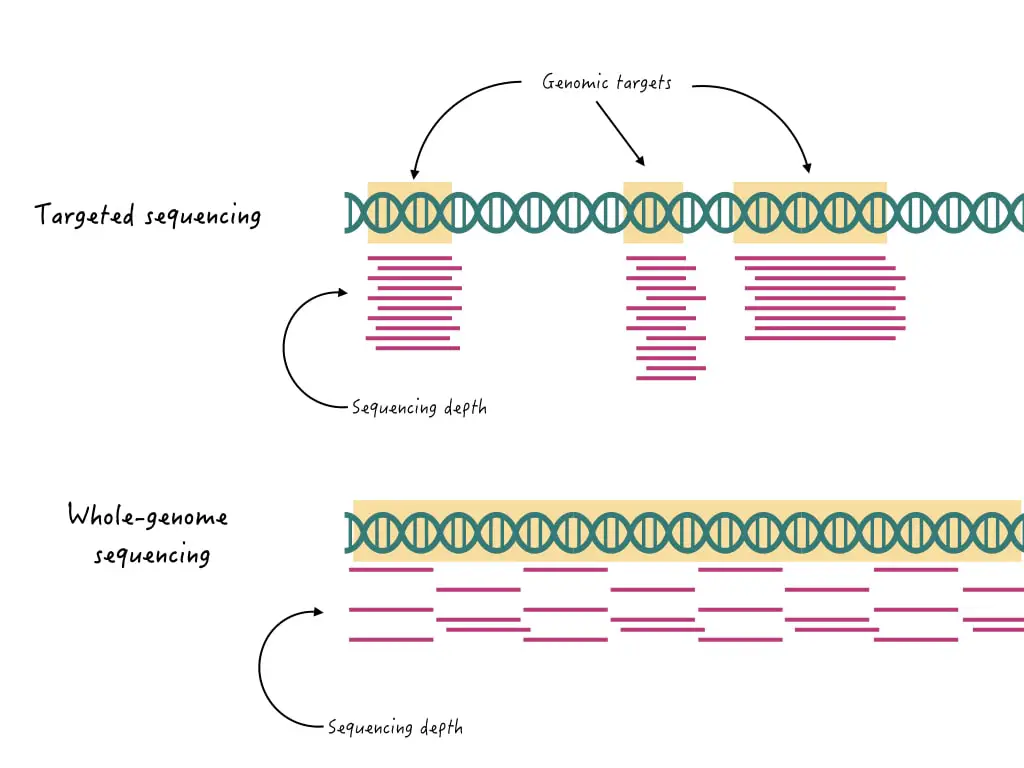
WGS has the power to sequence entire genomic content but lacks precision and depth, and can miss rare variants associated with the disease. Contrary, TS can be targeted to sequencing rare variants or associated genomic regions or genes with high depth and accuracy.
The shorter read format of WGS often misses crucial genetic information while the longer read length of the TS can accurately cover a gene or rare variants.
WGS is tedious, time-consuming and costly while TS is a simple, fast and cheaper technique.
WGS requires more computation and bioinformatic tools and software to manage, study and interpret the data. On the other hand, TS requires less complicated computation and bioinformatic tools.
Companies provide ready-to-use kits and software to analyze various genes, and variants thus the process is more streamlined and simple. In conclusion, TS is an important tool to study disease-associated rare genes and variants.
Keep in mind that targeted sequencing doesn’t mean targeting a single gene or sequence, many genes or sequences can be targeted in a single run. This benefit also helps to analyze every possible gene associated with a disease like cancer.
WGS can only run a single sample while TS sequencing can run multiple samples or an entire gene panel.
It is important to note that prior sequence, variant or gene information is required to perform TS as it dedicatedly studies only a single target (or many one after another).
How does it work?
Targeted sequencing is available in both Sanger and Next-generation sequencing platforms and uses the same working principle of each technique. Notedly, Sanger-based targeted sequencing can sequence a single or a few genes while NGS-based TS can sequence many genes.
The working principle of TS is similar to whole-genome sequencing with an extra step of target enrichment. Target enrichment is a process to generate copies of the target gene or sequence. PCR amplification and Hybrid capture are two types of target enrichment modules used in the TS.
Primer-extension method is often used in some sequencing techniques. Let’s see each method for target enrichment one after another.
PCR amplification:
In PCR-based target enrichment, a target region is amplified in the polymerase chain reaction before sequencing. The target-sequencing primer anneals with the target location and amplifies it with the help of Taq DNA polymerase.
Uniplex and multiplex PCR amplification are also available to simultaneously amplify one or various regions from the DNA, respectively. However, multiplexing has lower sensitivity and accuracy compared to the Uniplex reaction.
Reaction failure, false amplification and primer non-complementation are common errors in multiplex PCR reactions. After enrichment, the DNA sample is sent for sequencing. Reaction bias, inability to amplify large genomic regions, and false-positive amplification are common problems in PCR.
Note that prior sequencing information is required to design primers. Also, a known sequence tag is added to the sequence when dealing with multiple sequences or genes.
Hybrid capture:
In the hybrid capture, additional hybridization (biotinylated) probes specific to the sequence are added for enrichment. After successful hybridization, the rest of the genomic DNA and unbound probes are removed by washing.
Streptavidin-coated magnetic beads are added to attract the biotinylated probes hybridized with the DNA, the rest of the things are removed by washing.
TS by Sanger sequencing:
Sanger sequencing is a gold standard method for sequencing larger genomic regions. It has a read capacity of 100 bp which is pretty long and good for targeted sequencing. It also has excellent sequencing depth and precision.
However, unfortunately, it can’t sequence the entire gene panel. It’s suitable for one or a few targets or genes.
Related article: How to improve Sanger Sequencing Results?- 5 Technical Tips from Experts.
TS by NGS:
NGS (Next-generation sequencing) is an amazing tool to sequence many regions or genes in a single reaction with utmost precision. Using a short read technology, the hybrid capture enriched target is amplified in the bridge amplification reaction.
Kits are available for single genes, genes panel, or disease-related genes. In addition, handy analysis software is also available for data analysis.
(I think I do not have to explain Sanger or NGS, separately).
Advantages:
- More read depth and sequencing precision.
- Can sequence only target genomic regions.
- More manageable data sets and straightforward data analysis.
- Detection of rare genetic variants.
- Detection of low-frequency genetic variants.
- Higher resolution and coverage.
- Allow multiplexing and multiple sample sequencing.
- Cost-effective- the average cost of targeted sequencing is much lower than the WGS.
Limitations:
- The major limitation of TS is that it requires sequence information to design primers for PCR amplification and sequencing and thus can’t be utilized for de novo sequencing.
- Tedious probe design and hybridization procedure also make the entire procedure tedious.
- TS has a limited scope and can not sequence non-target regions effectively.
Applications:
TS is widely accepted in the field of disease study and diagnosis, cancer research and pharmacogenetic studies.
Targeted gene sequencing:
A recent study published in the Scientific Reports by Abu-Helalah et al. (2020) showed the importance of the TS for the analysis of particular genes. They sequenced the BRCA1 and BRCA2 genes from the high-risk Jordan breast cancer patients and reported 19.1% pathogenic mutations from both genes.
Notably, some of the reported variants were so rare. This study shows how accurately targeted gene sequencing can study genes and identify rare variants. Note that they have performed PCR-based target enrichment.
TS in cancer research:
Targeted sequencing is highly advantageous for cancer studies and research. Various rare and common genes can be investigated using various TS strategies to screen, diagnose and study cancer.
A study published in the Computational and Structural Biotechnology Journal by Bewicke-Copley et al. (2019) showed how targeted sequencing benefits the study of somatic, rare, and copy number variants. They explained the importance of TS for cancer research, postulated the procedure, and bioinformatic tools to extract meaningful information.
Another study published in Scientific Reports by McCabe et al. (2019) developed and validated target sequencing gene panels for various cancers. Their panel is capable enough to screen single nucleotide variations and indels from various types of cancer.
Pharmacogenomics and precise medicine:
TS has great value in cancer research and disease diagnosis, however, it can also be utilized in pharmacogenomics, precision medicine and personalized medicine fields too.
A study published in BMC Medical Genomics by Gulilat et al. (2019) emphasizes how important and valuable the tool TS is for precision medicine. They have developed a novel pharmacogenetic panel for CYP2D6 gene copy number variations using NGS-based targeted sequencing.
Accurate sequence and variance information has been collected using a short-read sequencing approach, in the present study.
HLA typing:
HLA typing is a procedure to match the HLA genetic print between donor and recipient for organ transplantation. Accurate typing nullifies the chances of graft rejection. TS-based HLA typing has been increasingly used for organ transplants.
It’s an NGS-based targeted sequencing approach in which the highly polymorphic HLA regions are precisely sequenced. Research published in BMC Bioinformatics by Ka et al (2017) used the HLAscan tool and NGS-based targeted sequencing for HLA typing.
Their findings showed 100% accuracy for HLA-A, -B, -C, -DQB1, and -DRB1 with >90% of sequence depth. This manifests that the TS along with HLAscan is a valuable, user-friendly, simple and cheaper technique for HLA typing and validation of transplantation compatibility.
Related articles:
- DNA Sequencing: History, Steps, Methods, Applications and Limitations.
- Sanger Sequencing vs PCR: Common and Technical Differences.
- How to Prepare DNA For Sequencing or NGS?
Wrapping up:
In conclusion, target sequencing enables scientists to study specific genes or sequences from the genome and identify variants or genes associated with diseases. It has several advantages including its cost-effectiveness, accuracy, speed and precision.
However, despite the requirement of prior sequence knowledge and limited scope, it is an important tool for genetic studies, disease diagnosis and cancer research. I hope you understand the concept. Comment below and let me know your call on this.
Abu-Helalah, M., Azab, B., Mubaidin, R. et al. BRCA1 and BRCA2 gene mutations among high-risk breast cancer patients in Jordan. Sci Rep 10, 17573 (2020). https://doi.org/10.1038/s41598-020-74250-2.
Lee, Seunggeung, et al. “Rare-variant association analysis: study designs and statistical tests.” American Journal of human genetics vol. 95,1 (2014): 5-23. doi:10.1016/j.ajhg.2014.06.009.
McCabe, M.J., Gauthier, ME.A., Chan, CL. et al. Development and validation of a targeted gene sequencing panel for application to disparate cancers. Sci Rep 9, 17052 (2019).
Gulilat, M., Lamb, T., Teft, W.A. et al. Targeted next-generation sequencing as a tool for precision medicine. BMC Med Genomics 12, 81 (2019). https://doi.org/10.1186/s12920-019-0527-2.
Ka, S., Lee, S., Hong, J. et al. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC Bioinformatics 18, 258 (2017). https://doi.org/10.1186/s12859-017-1671-3.