“PCR is one of the most versatile, robust and widely utilized techniques used in various fields of science starting from genomic science to microbiology and food industry. It is used in disease diagnosis, screening mutations and identifying pathogens.”
A technique relies on the principle of amplification known as polymerase chain reaction is used in biotechnology, microbiology, environmental science, medical science, dentistry, anthropology, food industry, animal, and plant research.
A PCR machine is a powerful and sophisticated instrument that amplifies DNA in vitro. The functionality of PCR is based on the heat. During various heat steps, DNA denatures, primer binds and new DNA forms. Three major steps of PCR are denaturation, annealing and extension.
We have covered an amazing article on PCR, its principle, procedure and other information therefore we are not discussing it here. Read the article here: The polymerase chain reaction.
In the present article, we only have discussed several applications of PCR and try to explain it.
Key Topics:
Applications of PCR:
Diagnosis of disease
One of the groundbreaking applications of PCR is in the diagnosis and screening of human diseases. The inherited, non-inherited and infectious diseases can be screened using the present method.
Either using PCR directly or combining it with other techniques such as restriction digestion, sequencing and blotting, various genetic disorders can be diagnosed.
Inherited genetic diseases:
Inherited diseases led by mutations in a gene. The genes are located on chromosomes and transmit to the offsprings. Thus, a mutation that occurred in a gene is also inherited to the consecutive generations, results in genetic abnormalities.
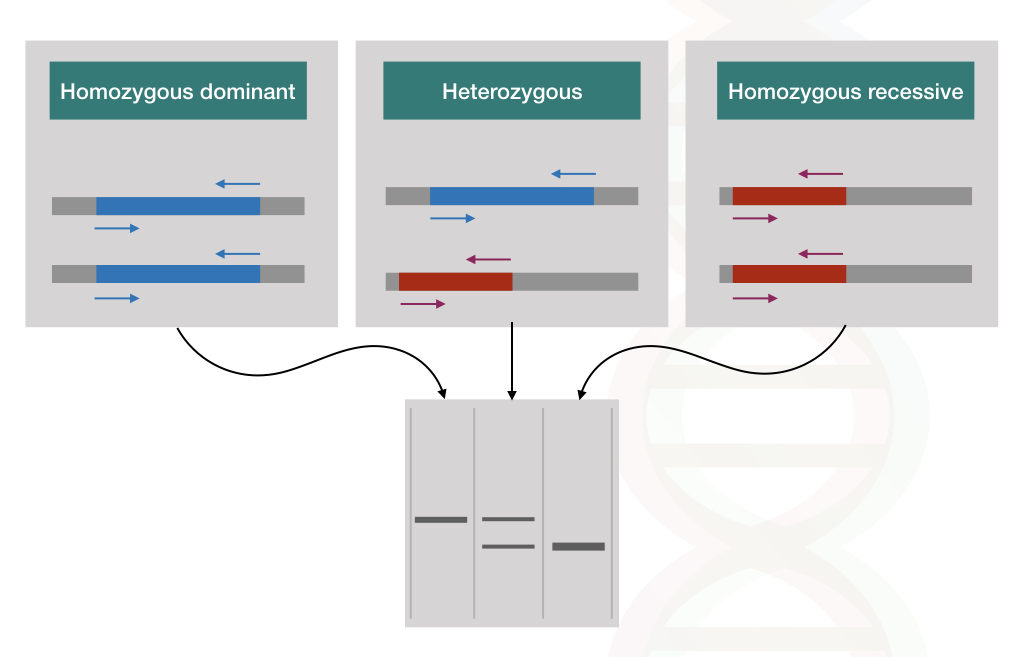
The condition might be autosomal dominant, autosomal recessive or X linked. Depending upon that, the individual may be a carrier of the disease (heterozygous) or affected with the disease (homozygous), Unlike other available methods, only PCR has the power to distinguish the homozygous from the heterozygous.
For example, thalassemia is an autosomal recessive disorder, only 25% of babies can be affected by the disease with two recessive alleles.
Using the PCR method called ARMS-PCR (Amplification refractory mutation system- PCR), two different sequence-specific primers amplify one mutant allele and one normal allele, respectively. Thus homozygous dominant without the disease, heterozygous carrier and homozygous recessive with two disease alleles can be distinguished on an agarose gel.
See the hypothetical representation in the figure below,
Using different PCR techniques such as Allelic specific PCR, touch down PCR, hot-start PCR and PCR multiplexing various genetic disorders can be identified.
Nowadays PCR assays for Huntington’s disease, cystic fibrosis, thalassemia, sickle cell anemia, Haemophilia, Myotonic dystrophy and Duchenne muscular dystrophy are available.
Read this series of articles:
non-inherited genetic disorders:
Cancer is a genetic abnormality arisen due to mutation or a group of mutations in some genes.
Mutations in genes related to cell differentiation and cell death commonly cause cancer. Genes help to control cell division and induce apoptosis are known as tumor suppressor genes.
Though cancer origins due to the genetic mutations, it is non-inherited (most cases), arise only due to the interaction of genes with the environment. Using realtime PCR assay, gene expression related to tumor suppressor genes or oncogenes can be estimated. (Some of the cancer are inherited).
Infectious disease:
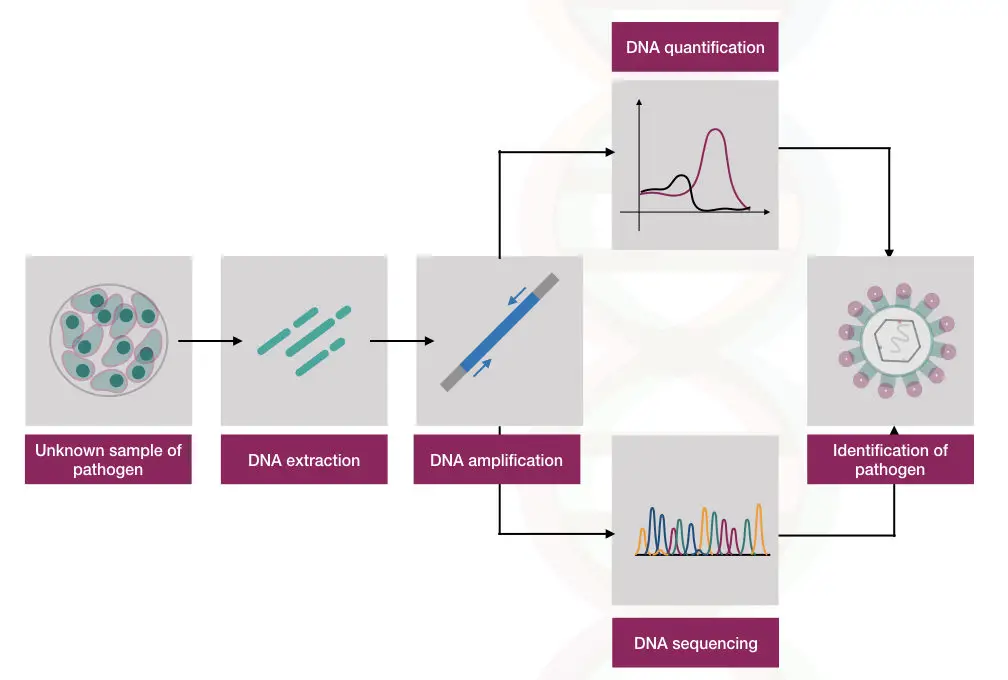
The real-time PCR assay is powerful enough to identify, characterize and detect pathogens, precisely. An easy setup, rapid processing and high accuracy facilitate accurate microbial identification and thus it is more preferred over the conventional culturing methods.
The present technique is used not only to identify microbes but also to quantify and diagnose various infectious diseases. It is a gold standard method for the diagnosis of tuberculosis called TB-PCR. The amount of the TB pathogen present in the sample can be quantified accurately within an hour.
HIV1-a causative agent of AIDS, Hepatitis B and C-causative pathogen of liver malignancy, cytomegalovirus- a causative pathogen of the immune inflammation and human papillomavirus a causative agent of cervical cancer are detected using the PCR. Furthermore, the amount of infection is also measured by mRNA expression analysis.
Selective DNA fragment isolation:
One of the basic primitive applications of the PCR is isolating the DNA fragment of our interest from the rest of the genomic DNA. To do restriction digestion, DNA sequencing or microarray, we need a high-quality DNA fragment of our interest.
Moreover, the total genomic DNA might be contaminated or not pure. The low quality DNA reduces the assay accuracy. Selective amplification allows one to amplify the DNA fragment, we wish to study. The amplicons are pure DNA products and used for downstream applications.
The amplification also allows an analysis of the DNA sample from the very small amount of starting material.
Microbiology: Amplification of microbial DNA
Traditional microbiology techniques are tedious and time-consuming to identify and characterize microorganisms.
Further, the chance of contamination and infection is also very high in those methods. Instead, the PCR amplification allows us to reduce time and increase the accuracy of results.
In a simple PCR set up, by utilizing a sequence-specific primer, any microbes or group of microbes can be recognized.
Also by doing DNA sequence any new microbial strain or pathogen can be detected. Yet another major advantage of the present method is the accuracy of results. The results of PCR are more accurate and reliable than conventional microbiology methods.
Related article: Microbial genetics: A rapid advancement in microbiology.
PCR revolutionalized the microbiology filed totally by removing the traditional microbial culturing. The hypothetical representation of how it is done is shown here,
Recent research:
The PCR is even used in the identification of different strains of one particular microorganism, for example, different strains of HPV-commonly found in cervical cancer, can be identified using different set of primers.
DNA fingerprinting
Using the PCR based genetic markers such as STRs ( short tandem repeats) and VNTR (variable numbers of tandem repeats) scientists can create a unique DNA fingerprint of any person. The STR and VNTRs are repetitive DNA sequences present in our genome with different repeat numbers.
(STR with 6 to 10 nucleotides long and VNTR with 5 to 50 nucleotides long).
No two individuals have the same VNTR or STR pattern in the world. Thus by amplifying either STR or VNTR or even both scientists can create a unique DNA pattern of different persons. Two individuals can be distinguished biologically and therefore it is so often used in criminal verification.
Criminal identification using the PCR based method accepted widely in every country for investigating a criminal case. Read our article on DNA fingerprinting.
Checking contamination
Checking contamination with PCR!! It sounds unrealistic at first glance. But PCR is one of the trusted techniques used in the checking of contamination and cross-contamination.
Let’s take an example,
Suppose we have a sample of HPV patients and it is supposed to be contaminated with some other microorganism.
This sample can’t be used for HPV culture, interestingly, using the sequence-specific primer of HPV Only HPV DNA is amplified in the PCR, any of the other DNA can’t.
And thus, in the end, we have the pure amplicons of only the HPV DNA only. These PCR amplicons are now can be used in DNA sequencing for further confirmation of results.
If we know the possible source of the contamination, contaminant strains of microbes can be identified as well using the sequence-specific primers.
In recent microbial practices, various economically important strains are regularly checked to detect contamination using the PCR.
Detecting Maternal cell contamination
One of the fascinating applications of PCR is in the checking of maternal cell contamination in the fetal sample.
The fetal samples of chorionic villi or amniotic fluid are taken for prenatal analysis for disorders like thalassemia, sickle cell anemia and cystic fibrosis.
During the fetal sample collection, the maternal cells or blood might contaminate the CVs or AF sample that leads to false-positive results.
As we discussed above, STR and VNTR patterns are unique property of a person, two persons are not genetically the same. Even not the mother and her baby. The contamination can be detected using DNA testing by PCR.
Sex determination
Sex determination can be done accurately using the PCR. A Y chromosome-specific marker is selected for it and amplified using a routine PCR protocol.
If amplification is observed, the fetus is male and if amplification is not observed the fetus is female. Moreover, some X chromosome-specific markers are also used.
PCR in Genotyping
A genotype is a genetic constitution related to a phenotype. Genotyping utilizing the PCR facilitates the identification and characterization of normal as well as the mutant alleles. Nested PCR, allele-specific PCR and other variations are used to do so.
Using two different sets of primers for two different genotypes homozygous normal, heterozygous, and homozygous mutant alleles can be identified.
This method is used in knock-out and knock-in mice genotyping studies in which the affected organism for different genotypes can be evaluated. However, the sequencing of PCR amplicons is highly recommended to confirm the results of PCR genotyping.
SNP (single nucleotide polymorphism) and SNV (single nucleotide variations) can be detected using the PCR genotyping method too.
See the image
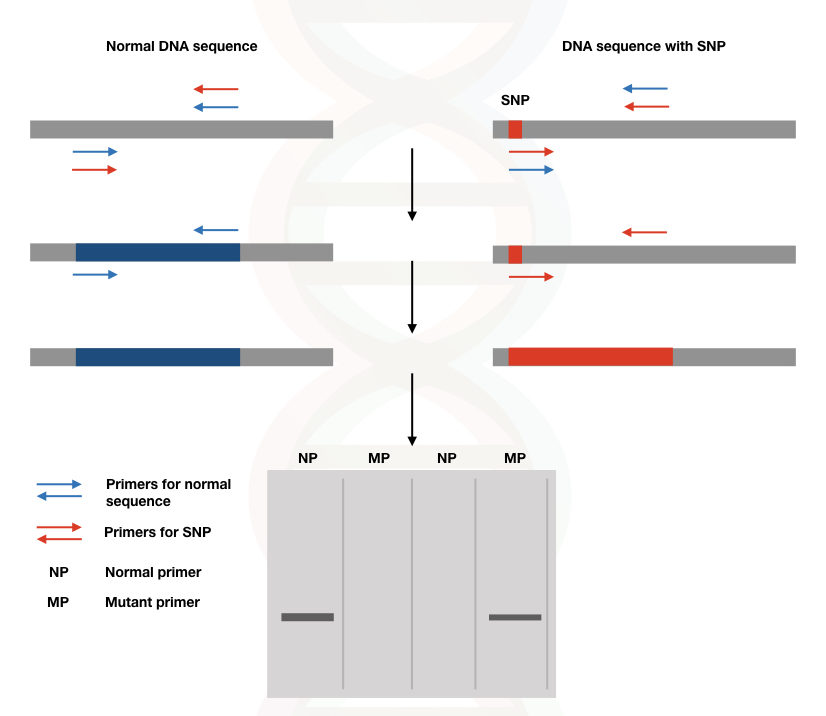
(Note: if DNA band is observed in NP and MP the individual is heterozygous for this particular SNP).
Read our article on SNP: An Introduction To Single Nucleotide Polymorphism (SNP).
Quantification of nucleic acid
Techniques like quantitative PCR and RT PCR (reverse transcriptase PCR) has the power to even estimate the amount of nucleic acid (DNA or RNA) present in a sample.
In the case of infectious disease, it is very important to measure the amount of infection, because the amount of pathogen-infection is directly proportional to the intensity of the disease or infection.
It measures each and every template DNA from every microbe present in a sample, ultimately we can determine the amount of infection. Quantitative PCR (also called Realtime PCR) facilitates the quantification of template DNA or RNA present in a sample.
If the sample is of retrovirus (having RNA as genetic material), by reverse transcribe it into DNA in a reverse transcription PCR, the amount of mRNA can be measured.
After each round of amplification, the amount of the accumulated DNA amplicons are measured by the real-time PCR machine. A fluorescently labeled probe or dye hybridize with template DNA, which emits fluorescent, detected by the detector.
The amount of the fluorescent emitted is directly proportional to the amount of the nucleic acid present in a sample.
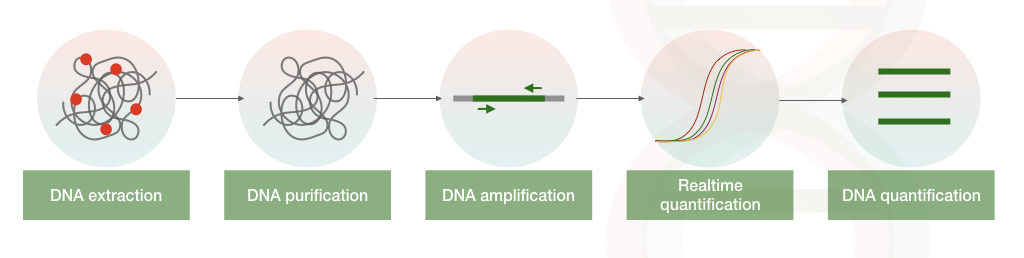
We have explained both the dye-based method and the fluorescent-probe based method is our previous article: Real-time PCR: Principle, Procedure, Advantages, Limitations and Applications.
Estimation of gene expression
The difference in the gene expression between cells, tissues or organisms can be examined using the PCR.
"When a protein is formed from a gene via the mRNA transcript is called gene expression."
Realtime (RT)-PCR is a gold standard method for routine estimation of cytokines and chemokines gene expression.
A cytokine is a component of the immune system that plays an important role during organ transplantation. Also involved in autoimmune disorders and inflammatory disorders.
By measuring the amount of mRNA or gene expression regulating cytokines; scientists can evaluate the effect of it and monitor its therapeutic effects in patients. The gene expression PCR assay is now a gold standard method used in clinical microbiology and oncology.
The quantitative PCR is further employed in gene expression studies in gene therapy experiments and RNA interference studies. In the RNA interference experiments, the amount of gene suppression or gene silencing is measured using the gene expression assays.
Moreover, in the gene therapy experiments it is used to measure the gene insert into plasmid as well as its expressions in cell lines.
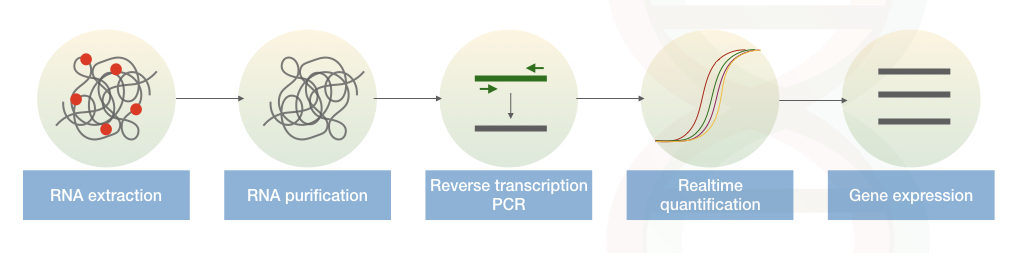
Note:
For the gene expression studies instead of DNA the RNA is isolated and it is reverse transcribed into the cDNA. This complementary DNA is amplified and measured in a PCR called reverse transcription PCR.
Validation of cell lines
Cell lines are developed for different in vitro gene therapy and gene transfer experiments. Those cell lines once cultured and can be validated using the polymerase chain reaction.
The cell from the cell line (possibly contains the insert DNA or DNA of our interest) is taken and in situ PCR is performed.
In the in situ PCR, the amplification is directly performed on the slide containing the cell lines.
Or
Instead of in situ PCR normal PCR can also be performed by using the extracted DNA from the cell line.
PCR cloning
PCR cloning is another mindblowing application of the PCR technique that allows DNA amplification and ligation in a single experiment, without doing restriction digestion. A rapid cloning method called direct PCR cloning allows researchers to clone DNA which is not available in a larger amount.
We will discuss the PCR cloning in some other article.
Plant research and agriculture
PCR in GM verification:
The genetically modified plant species are those whose genetic composition is changed, artificially in order to create new economically important traits. Some useful and beneficial traits and phenotypes are developed in plants by altering their genes or DNA.
Once the GM plant is developed, it is very necessary to verify it. To identify the insert its expression a PCR is used. Even, the method is widely accepted by consumers as well as developers who cross-check the quality of GM using the PCR.
Furthermore, the PCR is a gold standard method for grain handling and grain processing industries for certification and verification of grains. If you are interested in Genetically modified organism please read this article:
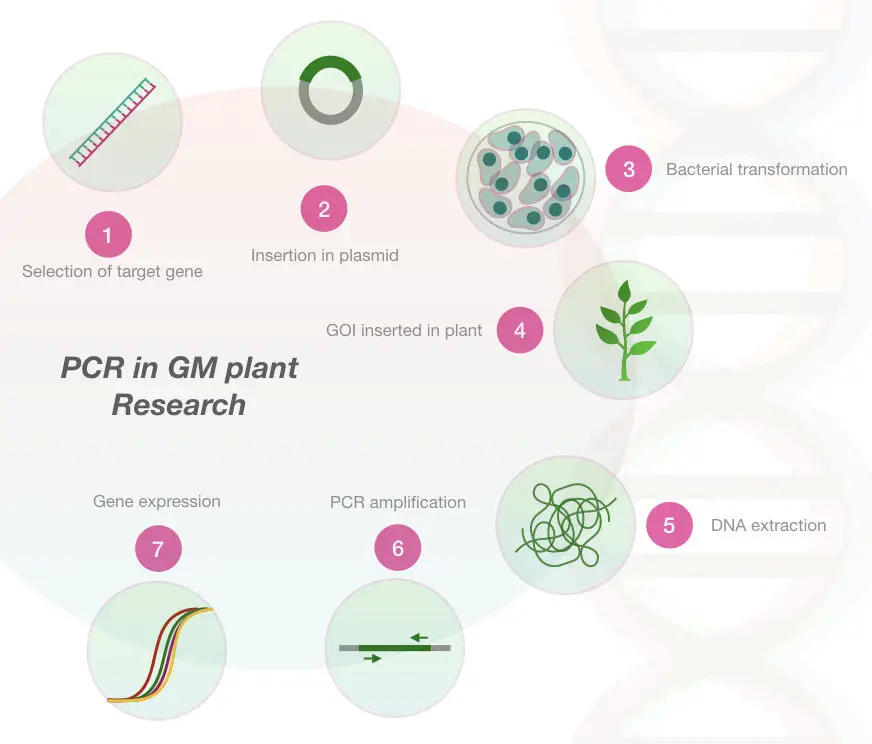
Marker assistance selection in plants:
Markers like RAPD (Rapid amplification of polymorphic DNA) identifies and characterize different plants and plant species whose base is the PCR. Thereupon, the phylogeny of plant species can be created using the PCR-RAPD based marker assistant selection method.
RAPD is a method in which the random universal primers are employed for amplification in PCR. Based on the sequence polymorphism and difference primers amplify DNA sequences and gives different banding pattern in agarose gel electrophoresis.
RAPD is simple and effective marker widely used in plant research.
Phyto-pathology:
Alike us plants also suffer from pathogenic infections. Plants may die with the infection and if it is economically important, one has to suffer from a loss. Viruses, bacteria and fungus are the groups of common pathogens infect plants.
Using the PCR detection method, infectious pathogens (for example Xanthomonas, pseudomonas, and mycoplasmas) for economically important plants are identified and subsequently, management steps are taken to prevent it.
Insert analysis:
For developing a novel economical important plant, scientists have to insert a desired gene into the plant genome. For that, a bacterial vector is used as a carrier of genetic material.
Using the PCR, the insertion of a gene into the vector is verified, in addition to this, the expression of that foreign gene in the plant genome can also be measured by the real-time PCR.
Apart from this, PCR and its variations have anonymous applications in plant research; some of them are:
- Sequence comparison of the marker gene (marker gene comparison studies)
- RAPD fingerprinting of different plant species
- DNA barcoding for different plant species
- Analysis of economically important proteins and its sequence.
- Analysis and comparisons of allozymes in plants.
PCR in animal research
We are using PCR not only in human and plant research and disease studies but also it is very important for animal research and disease studies. Inherited and infectious animal diseases are being diagnosed using the present method.
Some of the common applications of PCR in animal genetics are:
- In the identification of MTM mutated gene in dogs, responsible for X-linked Myotubular myopathy.
- For Bursal disease virus in avian samples.
- Identification of canine parvovirus in dogs.
- Deletion study of Meq gene in chickens
PCR in antiviral therapies
Antiviral therapies are now in trending, albeit, it is under the pre-clinical trial phase, not available fully for human subjects.
Scientists are now trying to cure AIDS, TB, and cervical cancer like infectious conditions using antiviral therapies by the HIV-1, HBV, HPV-1, and HSV-2.
The amount of viral nucleic acid reduced after the antiviral therapy dose is measured for checking the success rate of the therapy using the quantitative PCR.
PCR in dentistry
As we said above, PCR is now widely used in so many different fields because of its accuracy and reliability. Nowadays, it is used in dentistry and related fields in order to cure dental problems. S Ratti, S. mutants, S. sobrinus and S. ferus are common pathogens that affect dental health.
The sample is taken from the oral cavity, DNA is isolated and amplification is performed. The amount of pathogen present in the dental cavity can be measured. Furthermore, the realtime PCR like sophisticated methods are applied in the periodontal disease and endodontic infections.
PCR in artificial mutagenesis or site-directed mutagenesis:
Inserting a mutation in a DNA sequence (called artificial mutagenesis or site-directed mutagenesis) can be useful in removing restriction sites during gene transfer experiments.
The artificial mutagenesis can be done using different PCR based methods. two of the examples are given into the figure below,
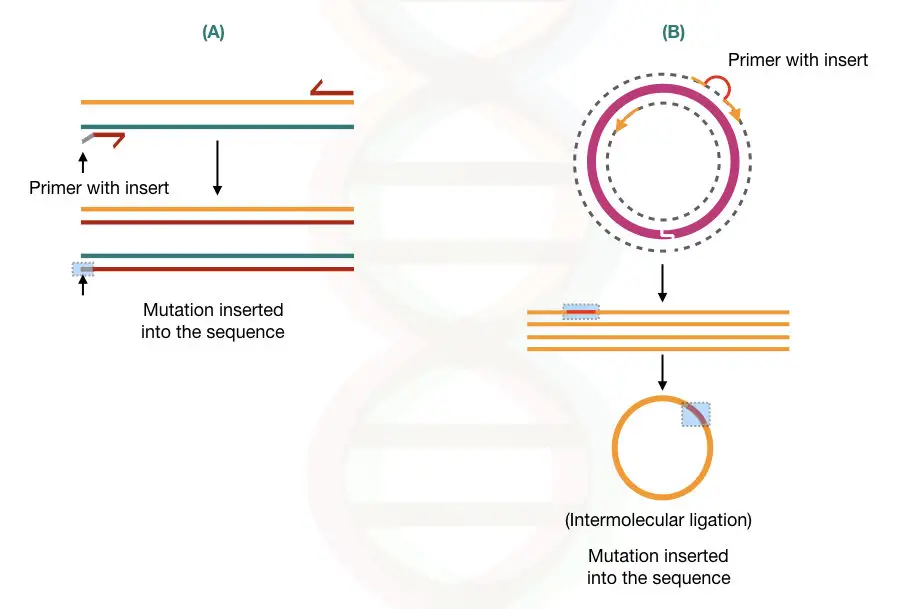
If you want to learn more about the present topic please read our previous article: Site-Directed Mutagenesis: Methods and Applications.
PCR in development of markers:
PCR is one of the basic technique which has replaced traditional cloning and blotting methods for the analysis of genes.
Gene analysis and study by cloning and blotting are replaced by the polymerase chain reaction. Therefore, PCR is used instead of tedious, time-consuming and inaccurate blotting, in the development of genetic markers. For instance, in RFLP, different sized DNA is amplified and identified in combination with restriction digestion.
Several PCR based markers are enlisted below,
- STS (short tandem repeat)
- VNTR (variable number of tandem repeat)
- SNP (single nucleotide polymorphism)
- STS ( sequence tag sites)
- SSCP (simple sequence conformation polymorphism)
- RAPD (random amplified polymorphic DNA)
- AFLP (amplified fragment length polymorphism).
PCR in DNA sequencing:
DNA sequencing is a method in which the nucleotide sequence is determined.
In fact, DNA sequencing is used for validating PCR results, once any of the amplicons is identified in the PCR-agarose gel electrophoresis. the results are cross-validated by DNA sequencing.
In DNA sequencing, a template or gene of our interest is amplified by a polymerase chain reaction to sequence it. So we can say PCR or amplification is one of the steps in DNA sequencing.
The template provided through the amplification is only the DNA fragment of our interest (which we wish to sequence) and also is contamination free.
Through the bridge amplification during the next-generation sequencing, the entire library of the DNA amplicons are being constructed. With all these a PCR amplicon assures efficiency, accuracy, and efficacy of results instead of using whole genomic DNA.
Related article: What is DNA sequencing?
Food pathogen detection using PCR:
PCR facilitates detection, identification and confirmation of food-borne pathogens and toxic fungus.
The risk of food contamination has increased due to the commercialization of food industries. Food processing, food packaging and food distributing industries are booming fastly. The packaged food reaches consumers between 3 to 4 days after cocking.
Although it is packaged under extreme sterile conditions, there might be some chance that the food product may be contaminated or infected by some pathogens. That pathogen creates hazardous effects on once’s health.
PCR is now widely used at different levels of the food distribution chain to detect possible contamination in it. The sequence-specific primers are used for most common food contaminating pathogens to amplify the DNA if any, in the food sample. If amplification obtained, the food product suspected to be contaminated with the pathogen.
The PCR based detection method is widely used in meat and meat-related industries. Furthermore, it is used in the dairy and poultry industries too.
PCR in the methylation assay:
DNA methylation is one of the epigenetic factors that have great impact on gene expression. A gene is deactivated once it methylated and vice verse. Thus to study gene expression, methylation study is required.
The amount of methylation for a gene of the DNA sequence is detected by using a special PCR modification known as methylation-specific PCR. However, the method for methylation assay is slightly different with an additional step of bisulfite treatment.
The bisulfite treatment converts the unmethylated cytosine into uracil but the methylated cytosine remains unaffected. A set of primers are designed, one specific for the methylated cytosine rich sequence with guanine. Another for the bisulfite-treated sequence in which the unmethylated cytosines converted into uracil.
This primer set has adenine in place of guanine which binds to uracil and then with the thymine in subsequent PCR cycles. The methylated and unmethylated sequence can be distinguished and quantified in the PCR.
Read more: What Is DNA Methylation?
PCR in cancer
The PCR is applicable in the diagnosis, prognosis, and monitoring of cancer using the realtime PCR assay. Even the MRD (minimal residual disease) can also be determined by the quantitative PCR.
Breast cancer, cervical cancer, chronic myeloid leukemia and other related cancer is detected using different PCR assays. Further, the amount of the mutant gene or oncogene can also be measured.
Interestingly, PCR is also used in cancer therapy monitoring. The success rate of the therapy, thus the expression of oncogene or mutant gene is measured each time after the therapy, using the quantitative PCR.
Read more on cancer: A Brief Introduction To Cancer Genetics.
PCR in gene therapy:
Gene therapies have a promising future. Scientists are now trying hard to make it real for all genetic abnormalities. The basic concept of gene therapy is to treat the faulty gene by replacing it or inserting a new gene in order to treat a genetic disorder.
The viral-vector mediated gene therapies have a more success rate in comparison with the non-viral vector-mediated gene therapies. PCR is used in the gene therapy for constructing a library for a gene of interest, to confirm the insert in a plasmid, checking transformation of a plasmid in cell lines and expression of GOI in a cell.
Read more on gene therapy: What is Gene Therapy? and How Does it Work?
Tracking animal migration using PCR
Now, this is something very innovative.
Environmental scientists use PCR very differently not for disease diagnosis, not for microbial identification but for tracking the migration pattern of animals.
They collect samples from the migration route of an animal, the sample might be a feces sample, urine sample, hair sample, blood sample or any other body sample.
In the next step, DNA extraction is performed from every sample. Using the species specif primer for that particular animal, the PCR is performed from the DNA sample collected.
In the final step, through agarose gel electrophoresis, the bands are matched to check if the sample is of the same animal or not!. If so, then based on data of sample collection site, the migration map of the animal is created.
(if the sample belongs to that animal PCR amplify it)
PCR in organ transplantation
Bone marrow transplantation is one of the therapeutic options for curing thalassemia and sickle cell anemia like disorders.
Kidney transplant and liver transplant practices are now common in medical science but in the very first step of organ transplantation checking graft-acceptance and graph-rejection is very necessary. If the graft is rejected, the transplanted organ can induce a strong immune response.
Hence, it is crucial to check the compatibility between donor and acceptor. PCR is the most-trusted method for checking the acceptance and rejection of the graft. Human leukocyte antigen locus present in our genome is highly variable regions that are used as a marker to check graft condition.
As like the DNA fingerprinting, the HLA typing is a unique genetic property of every individual. Different persons have different HLA patterns. If the HLA pattern matches between acceptor and donor and then the organ transplantation can be done.
A simple HLA locus-specific primers are used for doing the HLP typing PCR. If the locus is present, the primer will amplify otherwise remain unamplified. The same set of primers is used for both donor and acceptor to check similarities.
Library preparation:
PCR is used in the preparation of gDNA, cDNA or DNA fragments library. Using the adapter (known) DNA sequences, an entire library of DNA can be constructed and saved for future use.
Sequence tag site markers are created using the amplification-based library preparation and those markers are used to validate the library.
Construction of plasmid:
The plasmid is a single-stranded- circular bacterial DNA, used as a genetic engineering tool to transfer the gene of interest at the target location in a genome. The validating insert is also very critical to understand the experiments thus here also PCR is used to determine the insert whether it is inserted correctly or not.
DNA sequencing and polymerase chain reaction plays a critical role in gene therapy experiments. In addition to this, scientists also can detect other important sequences like antibiotic-resistant genes, the origin of replication, and marker DNA.
Read more on plasmid: Plasmid DNA- Structure, Function, Isolation And Applications.
Transcriptomics studies:
The transcriptome is an entire set of RNA or mRNA present in a cell or cell population. A novel modification of PCR, reverse transcription PCR is used in the transcriptomics studies for constructing the cDNA and quantification of mRNA present in a sample.
It is also used during the RNA-seq for cDNA library preparation. The cDNA is a complementary DNA reverse transcribed from the mRNA. Read more: What Is Transcriptomics?
Conclusion
Due to the high accuracy and amplification capacity, the PCR is an important tool in genetics and genomics research. It saves time for some critical experiments, which takes two to three days to complete, otherwise.
The PCR is applicable in almost all fields of science nowadays. However, it has several limitations that make it restricted to use. We will discuss its limitations in some other article.
Sources:
- Lenstra JA. The applications of the polymerase chain reaction in the life sciences. Cell Mol Biol (Noisy-le-grand). 1995;41(5):603-614.


